A Culture of [Blood] Cultures
Why hasn't rapid sequencing for serious infections and sepsis become standard of care?
In 2014, Sharon Peacock, a professor of microbiology at the University of Cambridge, wrote an important piece in Nature, first page shown below, advocating sequencing as a standard of care to rapidly determine the causative pathogen and direct appropriate therapy—since the sequence can reveal mutations that confer resistance to antibiotics. She wrote: “Microbial sequencing should be done as close to the patient as possible.” To achieve the best outcome, the need for speed to crack the case and initiate the right treatment cannot be emphasized enough.
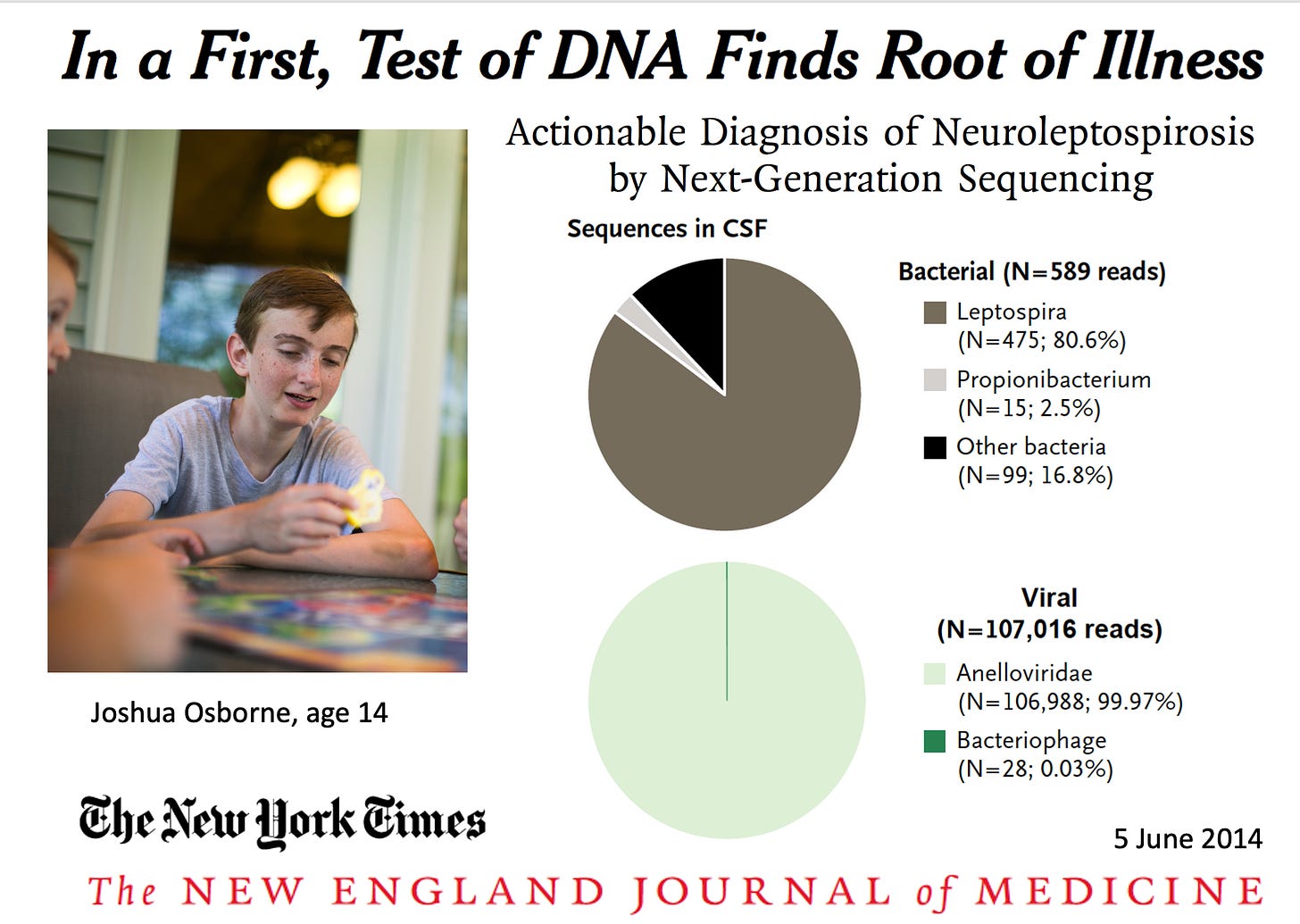
In that same year, 2014, we saw the life saved of Joshua Osborne, who had sequencing of his cerebrospinal fluid finally establish (within 48 hours) the elusive diagnosis of leptospirosis after many weeks of an extensive battery of tests, including a brain biopsy, that had failed to establish the cause of his encephalitis. About half of such infections of the brain are left undiagnosed with current approaches which include lab screening for a variety of pathogens and culture of the cerebrospinal fluid.
So let’s fast forward almost 9 years. What has changed to incorporate sequencing as standard of care for serious infections or sepsis? Essentially nothing.
Today when a patient presents with possible sepsis we draw multiple blood cultures and wait a few days before the results come back with a possible pathogen and readout for antibiotics that may be useful. The patient is bombarded with “empiric, broad spectrum antibiotics” to cover all the bacteria that are thought to be potentially causal, with implicit acknowledgement that viruses and other pathogens (fungi, parasites) won’t be covered by the antibiotic cocktail. That “blitzkrieg” cocktail, which may not even be directed to the underlying pathogen, has potential toxicity for the kidneys and other vital organs, and is typically continued until the cultures come back. All too often the cultures are negative, not revealing a/the pathogen, or show a contaminant, so, dependent on the patient’s clinical condition, the cocktail is continued for a full course of several days or longer. More potential for adverse effects of potentially misdirected antibiotics.
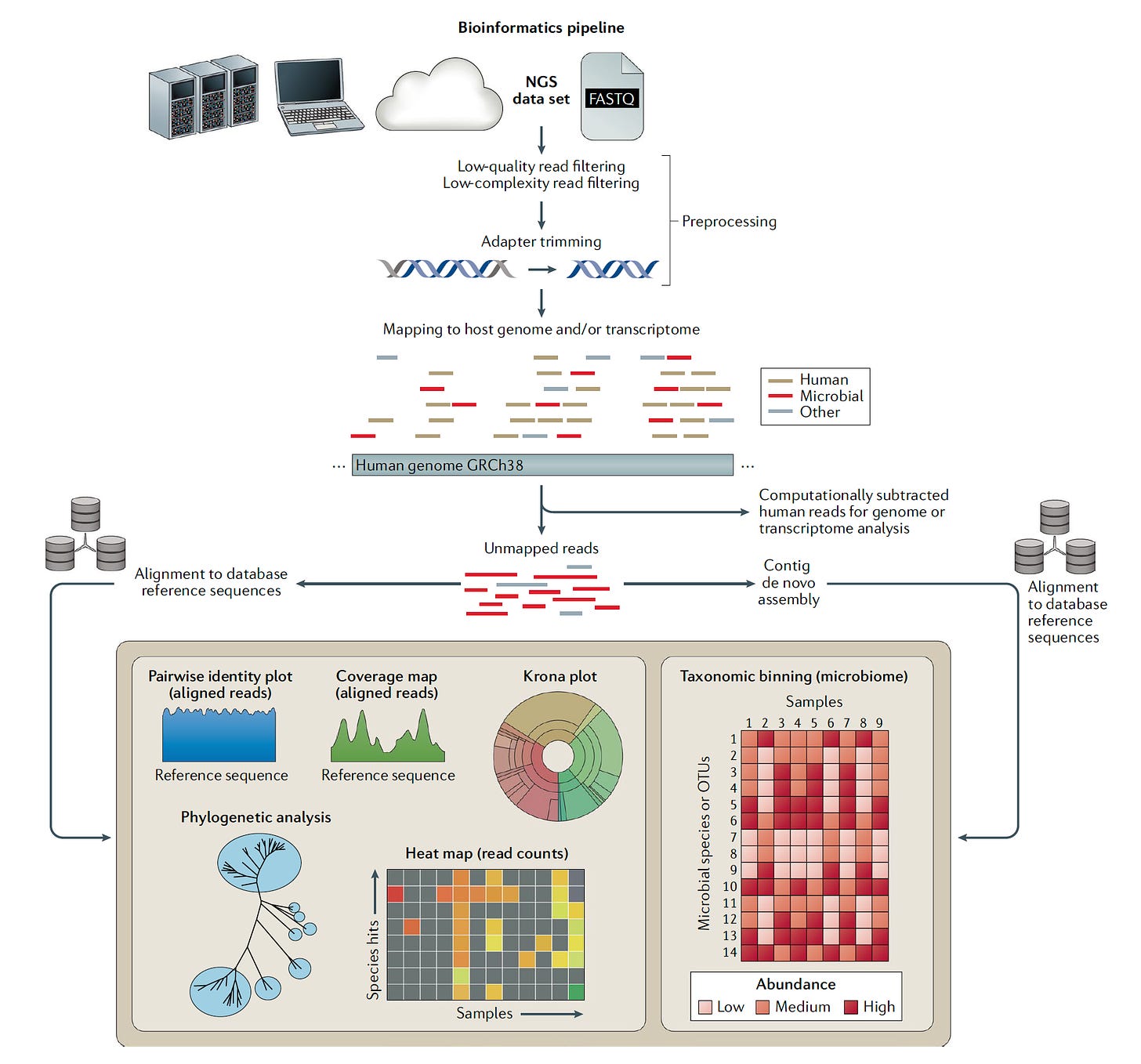
In contrast, “clinical metagenomics” as the field is known, takes an agnostic approach as to what is the causative pathogen. As shown in this excellent review, sequencing can be performed on any body fluid or tissue and there are several steps required, besides the sequencing, that include library preparation and bioinformatic analysis. There is no assumption about the bug—many cases successfully diagnosed by this technique would have been unthinkable pathogens, like an ameba, fungus or tuberculosis. The sequence tells the story in place of a clinician’s intuition for what might be the cause of infection.
Indeed, in the years since 2014, there have been many serious infections for individuals and outbreaks that were fully traced through sequencing, culminating in a 2019 New England Journal review stressing the importance of pathogen sequencing for public health.
Little did we know that within a few months of that review, genome sequencing of a pathogen was about to become a critical navigational guide for the pandemic, first providing the SARS-CoV-2 sequence to rapidly design vaccines within days, and then to provide continuous surveillance of the evolution of the virus over time and throughout the world. Such efforts concentrated in many academic labs and public health departments across the United States and throughout the world, have been remarkably instructive for the virus’s acquisition of new functional mutations, and our ability to differentiate major variants from the thousands that have appeared since late 2019. We even learned that our effectiveness of monoclonal antibody therapies were in many cases dependent upon the variant, but have yet to be able to incorporate that into medical practice. For example, recently the US FDA removed the approval of all the commercially available monoclonal antibodies because of anticipated resistance, even though some of the current wave infections (e.g. BA.5 variant) would still be sensitive to bebtelovimab. Furthermore, sequencing SARS-CoV-2 repetitively in people who are immunocompromised has demonstrated how the virus can go through hyper-accelerated evolution inside a person’s body, which, if spread to others, may have been the mechanism responsible for the initial rise of the Omicron BA.1 variant.
With all this time, background, and reinforcement during the pandemic in the past 3 years, why hasn’t metagenomics become routinely used across medical centers?
There are many reasons, but predominantly this is due to unwillingness of health systems to invest in getting this technology integrated to patient care. Some health systems, after a patient has not had a diagnosis for a protracted period of time, and is unresponsive to antimicrobials, will send out a sample to outside laboratories such as UCSF or commercial entities (ID by DNA, Karius, Hugo Biotechnology, Aperiomics, Day Zero Diagnostics, and others) that will turn around the sample in <48 hours. But there’s a delay imposed by shipping and a significant cost (>$1000) involved. This is often a “last ditch” approach, and even then the success for diagnosis is about 50%, a rate that will continue to increase as more pathogen sequencing is done and data are shared (which isn’t being done across the various companies and labs).
But the samples sent out are a rarity. And the reason for that is akin to the overall issue—lack of reimbursement (the big R word). For health systems to invest in desktop sequencers, reagents, training, dedicated personnel, and bioinformatic support, it’s a big barrier without reimbursement or incentive to do so. Of course, the incentive ought to be that’s the best way to care for patients, but that isn’t the working model. The rise of “pocket sequencers” and concept of pervasive sequencing capabilities to lower costs has been hyped for several years (“There's going to one of these things in everyone’s lab”) but their development has had little impact in clinical metagenomics to date.
The financials aren’t the only reason. As shown below, the analysis of the data can be quite tricky, separating out the pathogen sequence from bystanders and contaminants, that requires development of advanced, automated interpretation tools, something that would seem an ideal target for deep neural networks and generative AI. The investment in interpretation tools would be wise, since the more diagnoses that are made with accuracy, the more diagnoses will be made with accuracy (that’s not a typo repeat, that’s the auto-didactic loop). There are also regulatory issues, since clinical laboratories are high regulated to insure quality control, which further adds the to challenge for health systems to want to take this on.
While the opportunities here are extraordinary but the field is unmoved, UCSF, a pioneer in this movement, has already gone on beyond pathogen sequencing. They recently published the integrated use of metagenomics of the pathogen with host gene expression in critically ill patients, which identified 99% of confirmed sepsis cases, and the ability to separate out sepsis from non-sepsis etiologies.
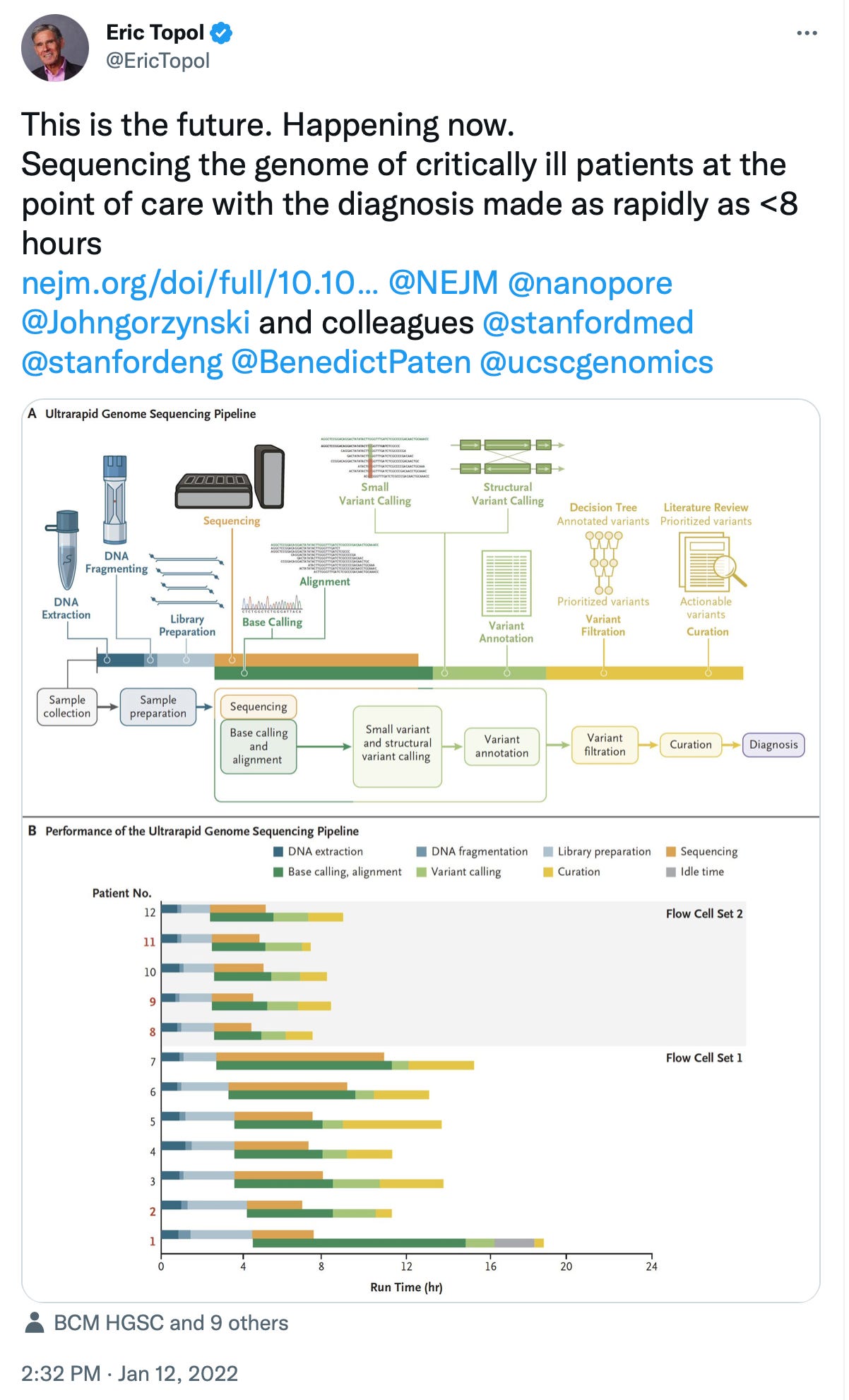
There has already been a published series of 34 patients in the intensive care unit in London where clinical metagenomics of the sputum within 8 hours led to the correct diagnosis with 92% sensitivity and 82% specificity, identifying unsuspected pathogens (such as Aspergillosis) and the anticipation of antibiotic resistance for bacterial infections. That workflow of 8 hours was also shown to be possible for whole genome sequencing of critically ill patients at Stanford University—not for infections but for undiagnosed illness via the host genome, a highly complex task. I posted about that earlier this year, a landmark achievement by Euan Ashley and colleagues.
One big reason we haven’t seen clinical metagenomics take off is our culture, which gets me back to the title of this post. We’ve been doing blood cultures for many decades (7 or more), and using broad-spectrum antibiotics (which have undoubtedly contributed to the crisis of antibiotic resistance) for many decades, and waiting for multiple days per patient for many decades. There’s considerable resistance to change in medicine, despite so many alluring aspects of this technology.
The bottom line: this can be done, but we’re not doing it. There appears to be no drive or incentive to change or adopt this technology for routine medical practice. Meanwhile, sepsis is a leading cause of death—it accounts for 1 of 5 deaths globally—about 11 million deaths in 2017. Patients and their families don’t know about this technology, but if they did, and started clamoring about it, we might begin to see some traction. I keep thinking of how many lives we could save (and could have saved) if only we responded to the charge in Peacock’s 2014 editorial.
Thanks for reading, subscribing and sharing Ground Truths!