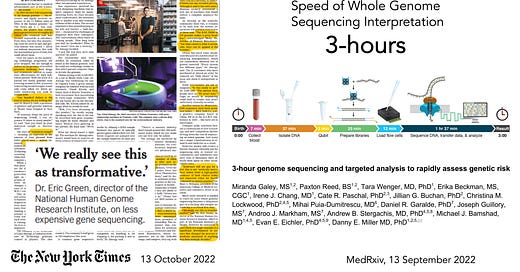
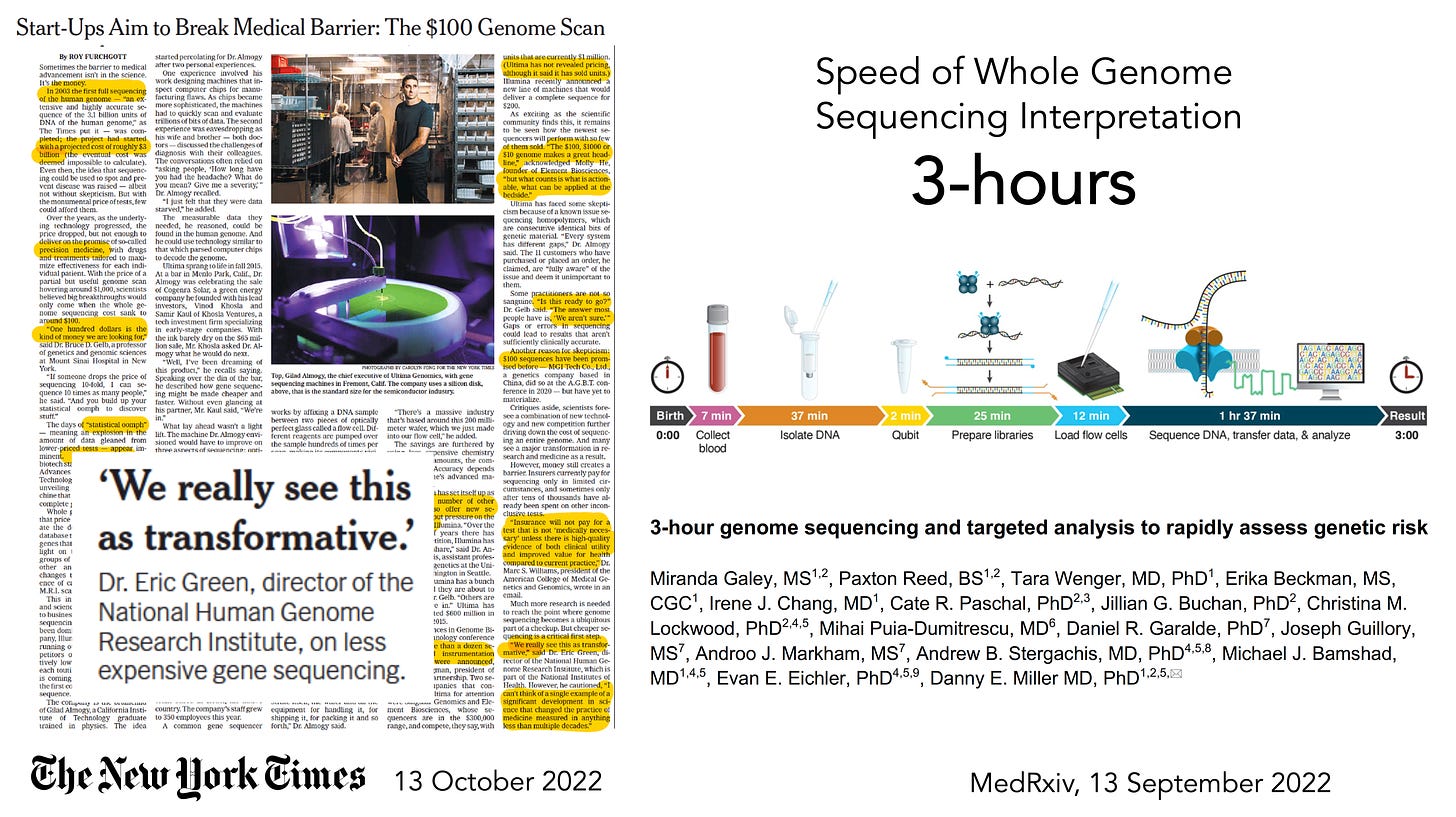
So much attention has been placed on the cost of whole genome sequencing (WGS) over the years, from about $300 million for the first one in 2000 (some estimates are as high as $3 billion), to now starting to approach $100. That’s a long sought and remarkable reduction in cost. But what is equally impressive is that a team at the University of Washington, led by Danny Miller, set a world record in September 2022, reducing the time from sample (at birth of a baby) to interpretation to 3 hours! That diagnosis (of lacking the pathogenic gene variant of concern) in a newborn was facilitated by knowledge of familial risk. Nevertheless, that acceleration of sequencing and analysis comes in the wake of the Stanford team, led by Euan Ashley, performing WGS in 12 people ranging from 3 months to 57 years, in a critical care setting, in as little as 7 hours and 18 minutes.
Stanford team rapid sequencing of 12 patients in critical care setting with molecular diagnosis made in 5 individuals, Table below.
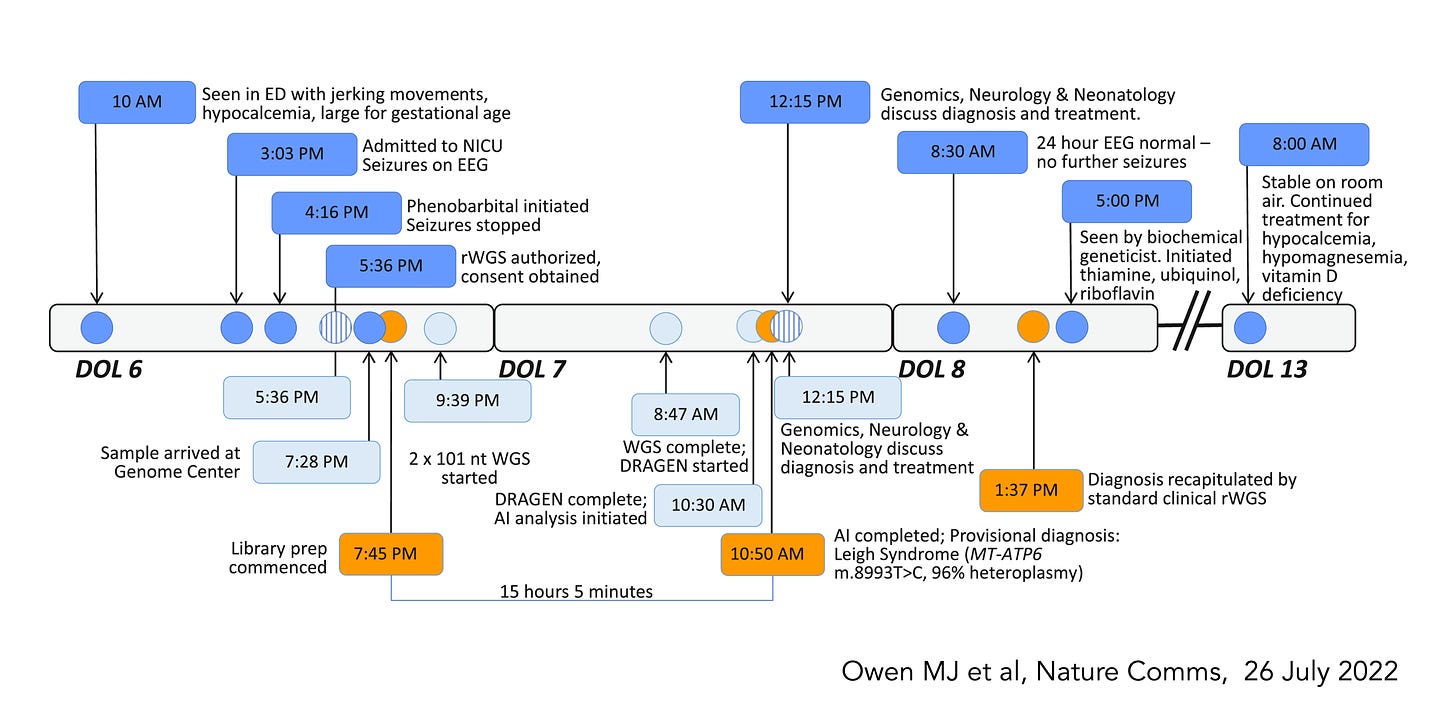
At Scripps Research, our SRTI team works closely with Rady Children’s Institute for Genomic Medicine, the group that has pioneered WGS in sick newborns who do not have a diagnosis, accomplishing this from sample to interpretation and management recommendations all within 13.5 hours, using multiple AI tools (labelled 1-3 below) to expedite the readout and care of the baby.
Here is a recently published example of a baby with successful treatment that was readily initiated after the WGS diagnosis of Leigh Syndrome was made.
That success story is one of several hundred with this program in sick newborns now operational in 83 children’s hospitals in the United States and Canada. Just think of the number of babies who have had irrevocable brain damage prevented or lives saved as a result of this initiative.
It’s unusual when there is a major innovation in medicine to start with neonates and children rather than adults. I actually can’t think of one offhand. Certainly WGS use in acutely ill patients has not yet been applied to adults, although elective use of WGS for diagnosing rare and unknown conditions has been occurring more at select academic centers in recent years.
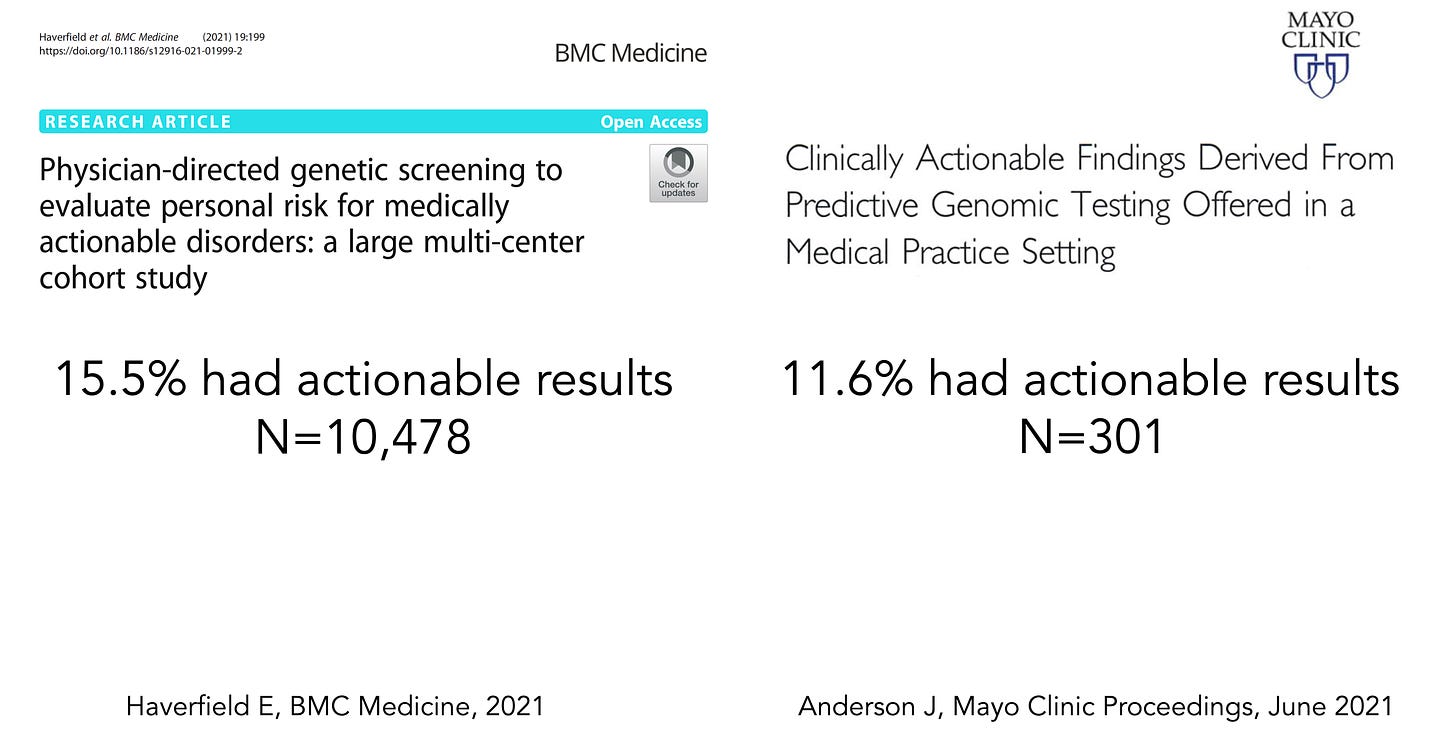
Now that WGS has gotten better (more complete and accurate), faster, and cheaper, will it become used more widely in adults? There are 2 fairly recent reports, here and here, that have shown that gene panels (sequencing just up to 147 genes) or WGS can be highly informative, picking up ~12-15% of healthy people with actionable results, such as important cancer or cardiovascular pathogenic mutations in individuals without a family history, or Factor V Leiden, posing a significant risk of clotting.
The >10% level is noteworthy. If we did a clinical trial with an intervention, such as a drug or device, and found that 1 in 10 patients derived important benefit, that level of absolute benefit would be declared as exceptionally important. Most randomized trials that are considered positive come with far less magnitude of benefit. The largest clinical trial that I led, with over 41,000 patients, demonstrated a survival benefit for 1 of 100 patients. Another frame of reference would be the Covid vaccine trials, which showed an absolute reduction of 6 per 100 (or 0.6 per 10) for preventing symptomatic infections out to 2 months (relative reduction, efficacy, 95%).
To also point out, the potential for WGS is far more than just picking up pathogenic mutations, such as a person’s gene-drug interactions, carrier state, and polygenic risk for most common medical conditions (heart disease, cancer, diabetes, etc). No less, there is no use of WGS yet in the acute setting, as we’ve seen in newborns, to help make the diagnosis when it is elusive.
The reduction in cost and time for whole genome sequencing is historic and one of the most important advances that has occurred in life science in recent years. With the increasing use of AI tools to make the variant calling and interpretation more accurate and rapid, along with contextualizing the medical literature for a molecular diagnosis and possible treatment, this could become someday an exemplar, beyond prediction of protein folding from amino acid sequence (AlphaFold), for AI’s contribution to biomedicine. Hopefully some day we will harness its value to advance individualized medicine.