We’re now well over 20 years since the first human genome was sequenced, but with few exceptions the massive amount of data that has been generated has not been transformed to routine patient care.
Over 30 million people have had their genome sequenced (exome or whole genome). The NIH All of Us research program released 100,000 whole genome sequences in March. (Disclosure: I am an investigator in the All of Us Research Program and an advisor to Illumina). The emphasis here is on research programs, since there has been very little use of whole genome sequencing in clinical practice.
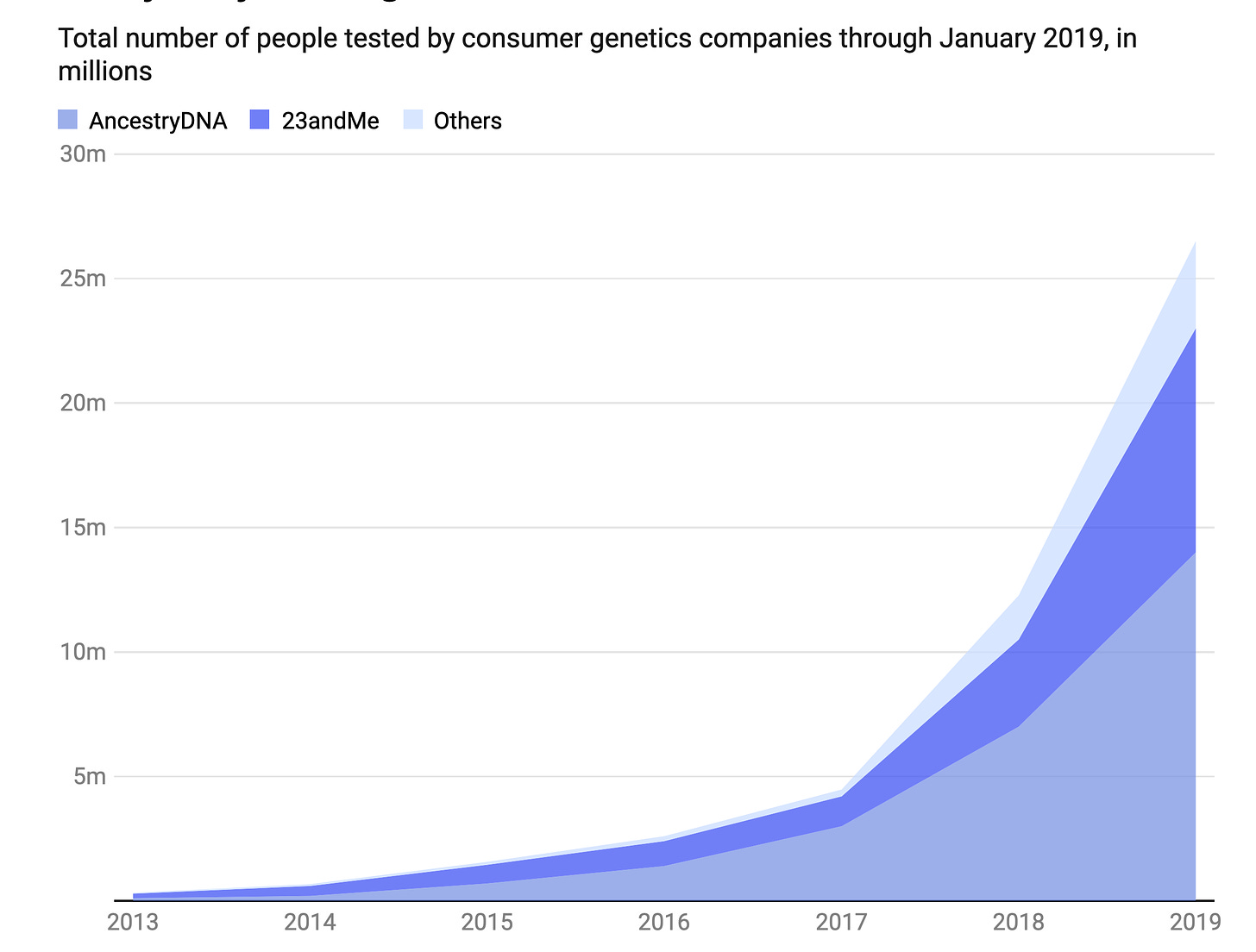
Well short of a genome sequence is a single nucleotide polymorphism (SNP) array, which assesses (genotypes) about 1 million letters of the 3 billion letter genome. These SNPs are common polymorphisms (those found in generally >5% of the population) that are associated with specific conditions. By early 2019, over 26 million people had a SNP array . Still today, many people think the SNP-array they had was a genome sequence, which is far less than 0.1% of what’s in a whole genome dataset.
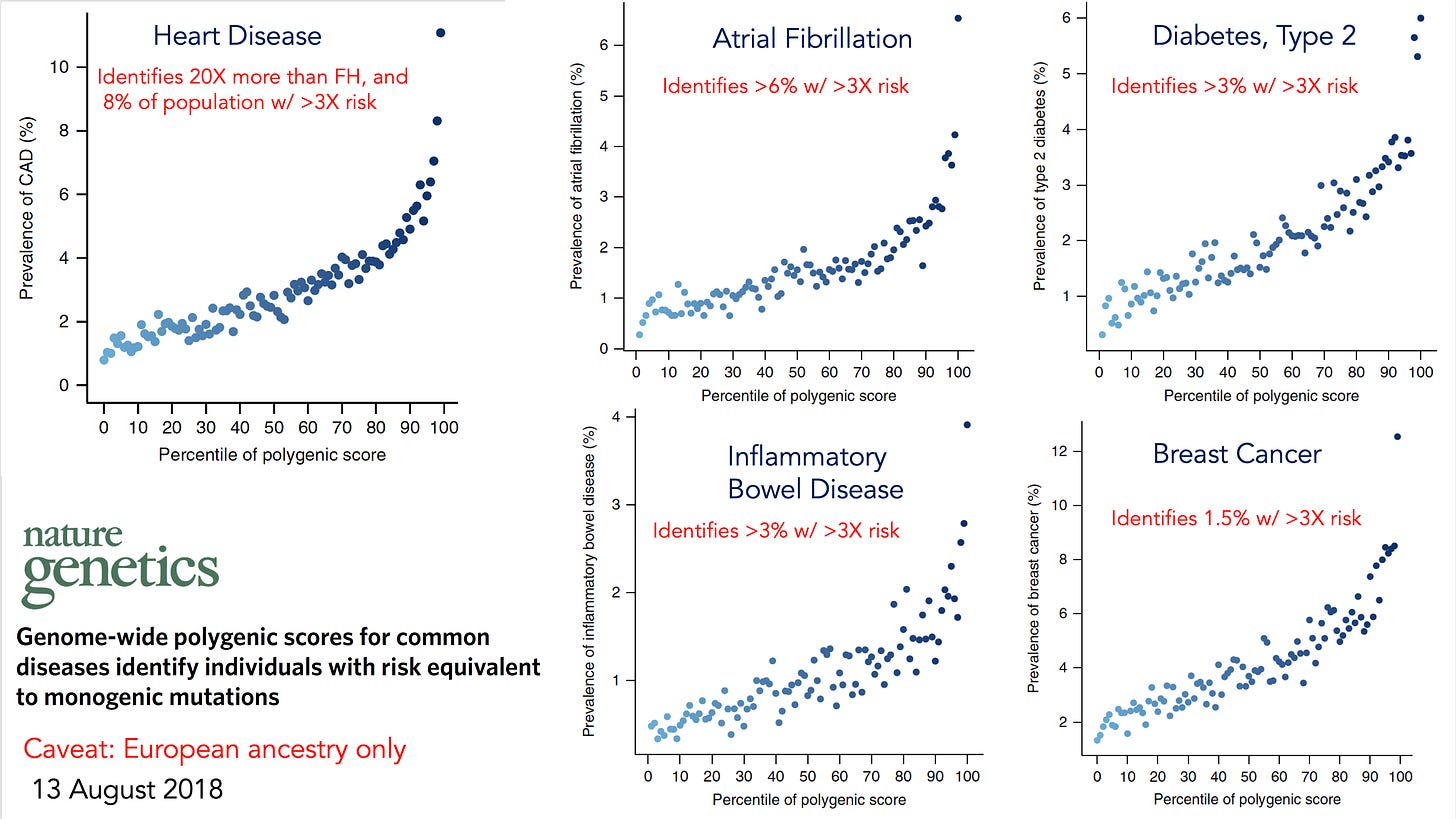
But the SNP-arrays can be used to determine polygenic risk scores for a wide variety of important and common conditions, including coronary heart disease, atrial fibrillation, breast cancer, colon cancer, prostate cancer, type 2 diabetes, inflammatory bowel disease, and many more.
By 2018 all of these had been extensively validated in people of European ancestry, and since that time most had expansion and validation across all ancestries.
The polygenic risk score identified people at high risk for each of these conditions, such as the 8% of the population with a greater than 3-fold risk of heart disease.
Editorials in leading medical journals in 2019 made some projections about the role of polygenic risk scores in clinical practice, as shown below.
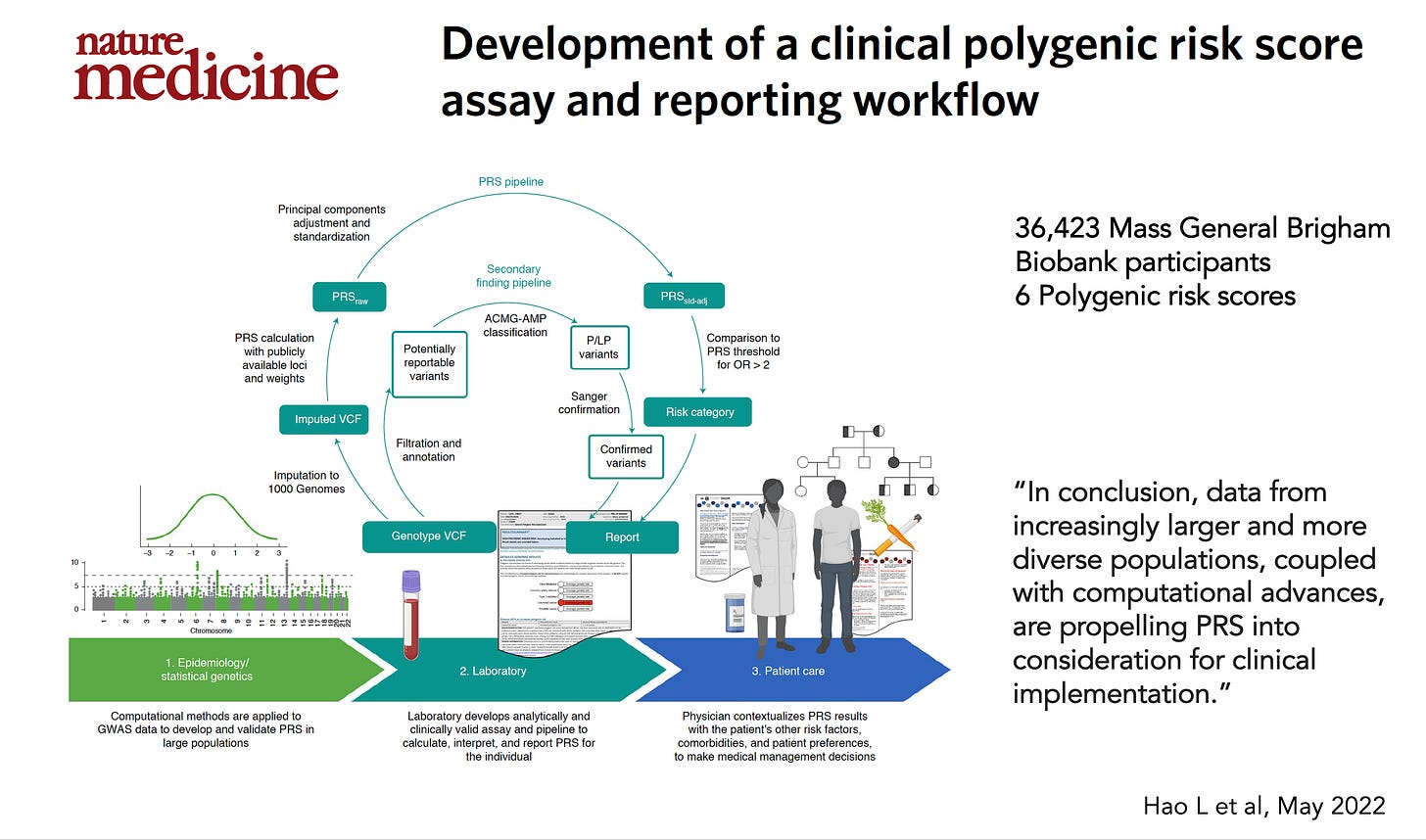
Yet more than 3 years later nothing has happened. The only significant report about clinical implementation was from Mass General Brigham that demonstrated feasibility
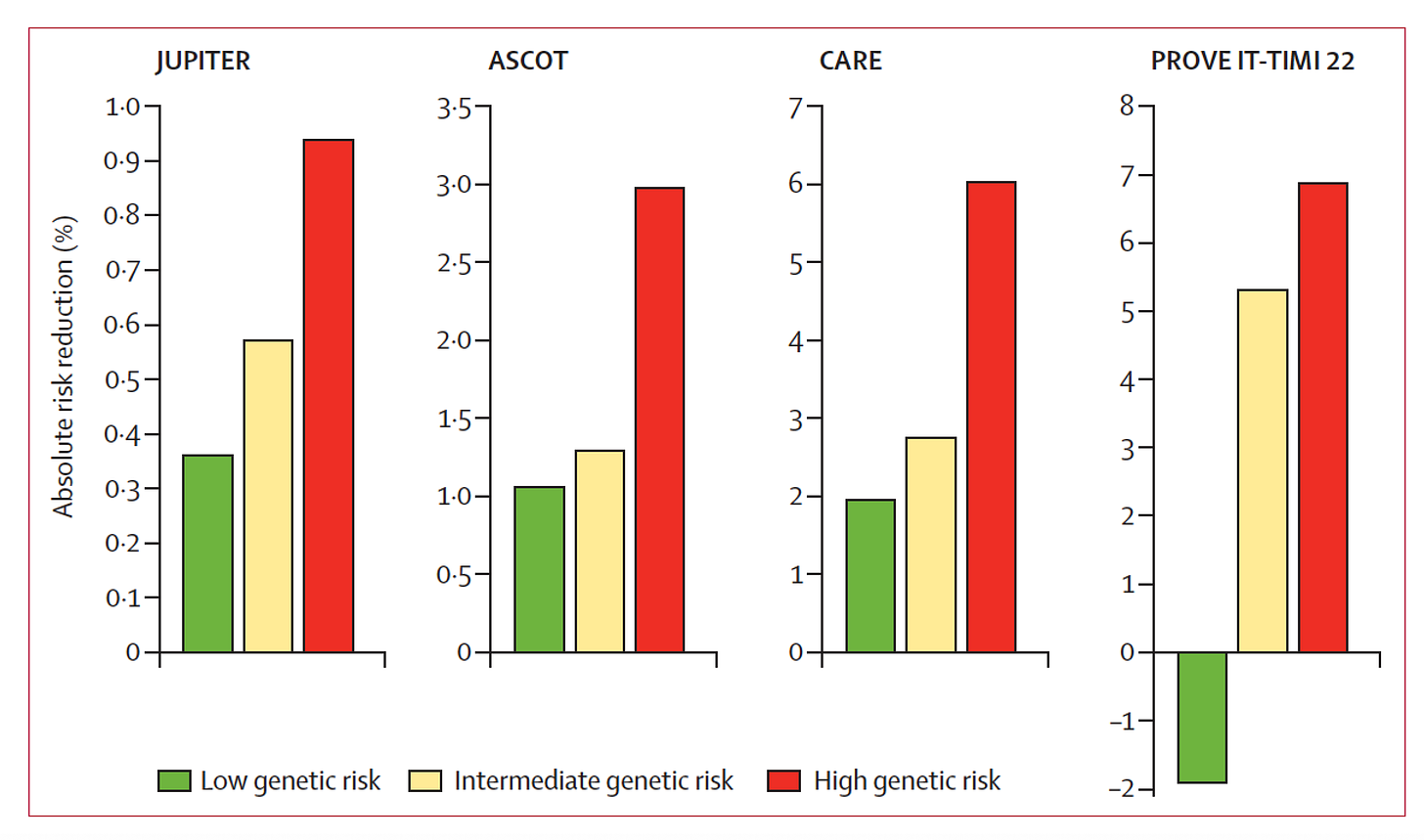
Polygenic risk scores can provide information beyond risk that is actionable. In the case of heart disease, helping the decision for the use of statins has been well established for many years (Figure below), and I’ve used that in my cardiology practice in real time, during a clinic visit, for patients who have had a SNP-array using the My Gene Rank app our team developed.
Now the SNP-arrays from 23andMe and Ancestry DNA cost $99, but they don’t provide polygenic risk scores which provide an independent and additive window into risk for a condition beyond the established clinical risk factors.
Meanwhile, a whole genome sequence had been steadily coming down in price, now at approximately $400-600, and with an announcement in June by one company at $100, the same cost of a SNP-array that provides >99.9% less data. There is skepticism that the Ultima company $100 genome is accurate or real, but it’s certainly anticipated that the cost of deep, high accuracy, whole genome sequencing will approach that level in the next couple of years.
Like many of you, I’ve had a whole genome sequence and found it to be of limited value. Why? Because the data assessed is not provided in a highly informative, user-friendly way. I want to see my major categories of meaningful data on my phone, which includes pharmacogenetics, polygenic risk scores, rare pathogenic variants, and carrier states. If I get a new prescription, I’d be able to quickly and simply review the data, share it with a pharmacist, to decide about my gene-drug potential interactions. These categories could easily be expanded. For example, we know a great deal about cancer predisposition genes from whole genome sequencing, but such data is not available. It sits in top-tier research publications.
That exemplifies the problem. For decades, the genomics research community has been in high gear productivity, extensively publishing data about medical conditions. But so little has reached clinical practice outside of a few exceptions like rapid neonatal sequencing or adults with serious, undiagnosed conditions, or in specific use cases in cancer. Even those are only performed by a limited number of health systems.
At least at this point, the “genome revolution” as proclaimed frequently and in the Nature cover (at top of this post), really seems to be confined to the research domain. The bottleneck in getting this into mainstream medicine used to be considered cost, but that excuse is likely to become untenable with many routine lab tests and scans at higher cost and potentially less informative. There is the problem of paternalism—the research community is afraid of “letting go” —believing the public is incapable of dealing with genomic data, such as variants of uncertain significance and the issues of probabilistic vs deterministic meaning of variants. Beyond these issues, the medical community has little grounding in human genomics. There are very few medical geneticists and genetic counselors relative to the population, about 1,500 and 6,000, respectively, for >330 million Americans. Education for all clinicians is vital but there has been no commitment or successful initiative at scale. This is unlike the National Health Service in the United Kingdom, the country leading the world in genomics, which has a major division known as Health Education England, that is responsible for educating clinicians about genomics.
During the pandemic we saw the power of pathogen genomic sequencing to detect major SARS-CoV-2 variants and track the virus spatiotemporally through the world, no less to do so in specific outbreaks. The relationship between the variants and specific treatments with efficacy (or lack thereof) for potentially lifesaving monoclonal antibodies was firmly established. Perhaps this will help facilitate the acceptance of medical genomics in the future.
The miscue that our genome sequence is our “operating instructions” must also be addressed, fully acknowledging that DNA sequence represents just 1-layer depicting human uniqueness, and does not by itself reveal the depth of information derived from all the other layers that include the transcriptome, proteome, epigenome, microbiome, immunome, physiome, anatome, and exposome (environment). Many of these layers are cell and tissue-specific (transcriptome, epigenome) or site-specific (microbiome), further emphasizing the complexity.
There remains vast potential to incorporate human genomics into medical practice. It is part of the ultimate vision for preventing medical conditions that individuals would otherwise be predisposed to manifest. For this to occur, all of the issues raised here, and more, will need to be addressed. Otherwise, we’ll essentially be stuck in 2 different orbits—one derived from the engine perpetually generating so much research, and the other—patients not deriving the benefit from all this extraordinary work.
Addendum
I’ve received feedback from some people and responses on social media that appear to have misinterpreted this post. There’s no question that genomics has had a profound and fundamental impact for unraveling genes and biologic pathways, across monogenic and polygenic diseases. Cancer is a genomic disease and it’s incontrovertible that a substantial number of patients have reaped the benefit of gene panels and tumor sequencing to define optimal therapies. Thanks to sequencing, people with undiagnosed, serious, rare diseases have had a molecular diagnosis made, which in many cases has been truly life-saving. I’ve written extensively about all that remarkable progress in 3 books along with multiple original research papers and reviews.
What I am referring to here is routine medical practice. Polygenic risk scores are not being used. Of the 30+ million of people who have undergone sequencing for research biobank purposes, how many individuals have had their results returned? Medical sequencing with return of results was initiated by one large health system —Geisinger Health (“My Code” program) in 2014. As of May 2022, 185,000 patients had exome sequencing and clinically actionable results (rare pathogenic variants) were returned to 3,400 individuals. (FH, BRCA 1,2, Lynch syndrome, Long QT syndrome/ related arrhythmogenic channelopathies, and some cardiomyopathies were the principal pathogenic variants detected). Geisinger reported that 3-4% of their patients sequenced had actionable results—that’s just from rare pathogenic variants. I’m not aware of other American health systems initiating large-scale sequencing efforts with return of results despite Geisinger’s efforts, which were tied to a multi-year contract with Regeneron. Beyond the factors that I’ve offered in the post, the lack of adoption of genome sequencing in medical practice may relate to absence of randomized trials or what would be considered compelling proof of improving major clinical outcomes. Whatever the reason, it hasn’t happened yet, despite multiple forecasts that we’d be well into the medical sequencing era by now. That may well happen in the future, but we ought to take a hard look at why genomics in general (outside of their use in a subset of patients with cancer, rare/undiagnosed diseases, and some pioneering neonatal programs) isn’t being incorporated to patient care at the individual level.