Medical Forecasting
The exciting opportunity to provide highly accurate risk and timing assessment at the individual level for actionable conditions to potentiate prevention
The following essay was published by me at Science today and I have adapted it for Ground Truths with additional graphics and new reports from this week.
“AI-Powered Forecasting” was recently on the cover of Science, highlighting a new deep learning model for much faster and more accurate weather forecasting (A morphed version of that cover for the current topic is found below). Known as GraphCast, it outperformed the gold-standard system and had an accuracy of 99.7% for tropospheric predictions, the most important forecasting region that is closest to Earth’s surface. Better warnings for extreme weather events such as hurricanes and cyclones will help save lives. The parallel in medicine is forecasting specific, actionable, high risk for individuals to prevent diseases or severe acute events. But we don’t have a gold standard for predicting health outcomes. That is hopefully about to change.
To frame where we are now, take the example of screening for cancer. Nearly 90% of women will never develop breast cancer, but current guidelines from the American Cancer Society are annual or biennial mammograms for all women aged 45 years and older. Using age as the basis for mass periodic screening for multiple types of cancer leads to the diagnosis of only 14% of all cancers in the United States. The main enrollment criterion for the largest clinical trial of a multicancer early detection test was 50 years of age or older, but a cancer signal was found in only 92 of 6621 (1.4%) participants, and only 35 (0.5%) were true positives. Now the Alzheimer’s Association has proposed that healthy people without symptoms should have a blood test for biomarkers, such as p-Tau-181 or p-Tau-217, that indicate β-amyloid accumulation in the brain and classification of “Alzheimer’s Stage 1.” In an era in which big, deep, and longitudinal data are available, relying on a simple, singular measure, such as a person’s age or the result of a blood test, to define risk is simplistic. It is a recipe for generating false-positive results, inciting alarm in people, and can be profoundly wasteful.
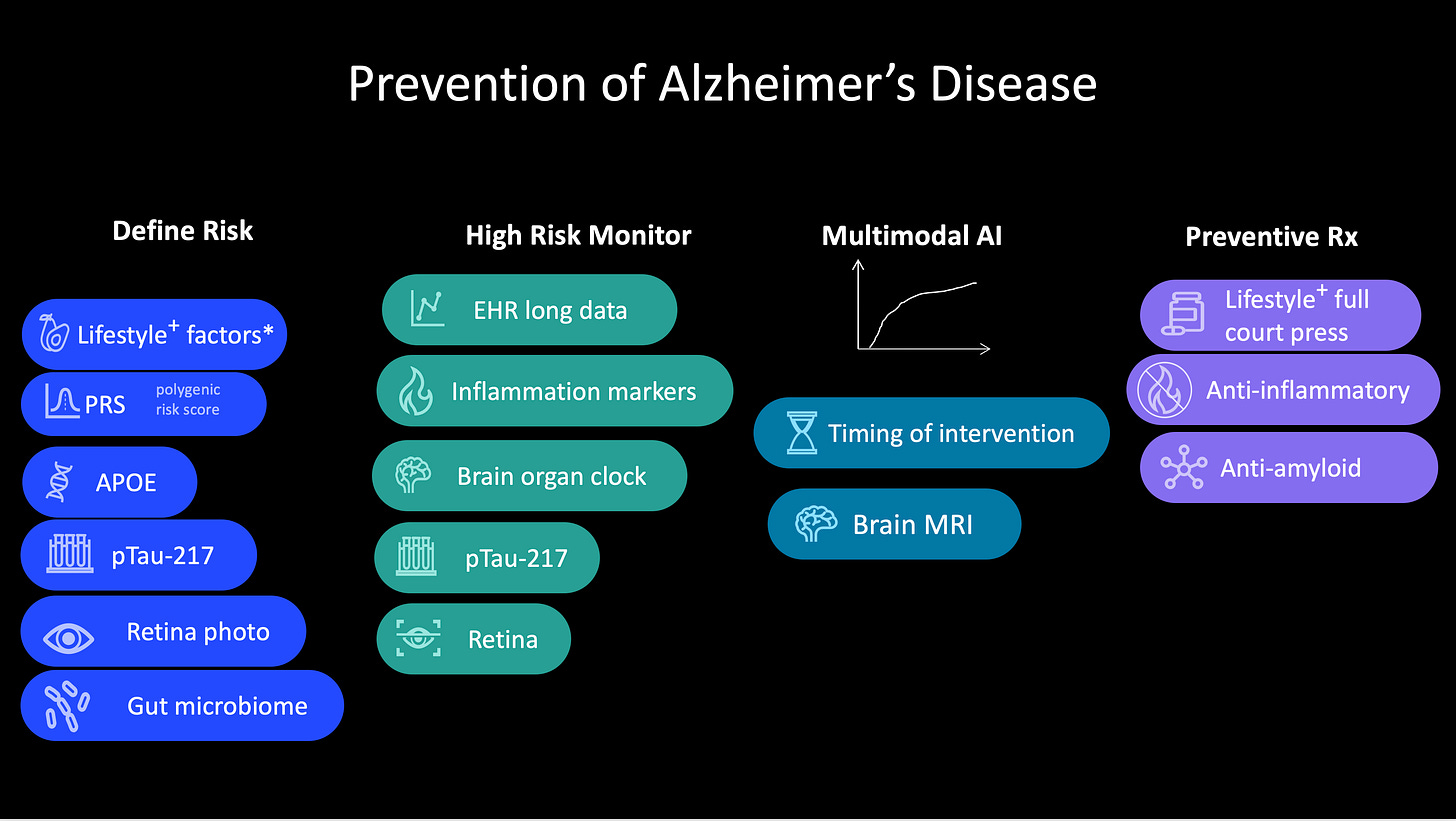
With multimodal AI, there’s an extraordinary opportunity to do for medicine what GraphCast did for weather forecasting (Figure below). Take the example of risk for Alzheimer’s disease, where there has been progress for blood biomarkers, but that’s just one layer of data. The risk of an individual could be assessed more accurately with orthogonal genomic data that include the apolipoprotein ε4 (APOε4) allele present in about 20% of the population, and a polygenic risk score, which provides complementary, independent predictive value. So can retinal imaging. Add to that electronic health records with both structured and unstructured text, imaging, and lab test results. Ideally, details of sleep history and physical activity would also be included but could otherwise be available from an individual’s wearable biosensor. A recent study using machine learning was able to predict Alzheimer’s disease up to 7 years before diagnosis by integrating electronic health record data for cholesterol, blood pressure, vitamin D, and sex-specific features such as osteoporosis in women or erectile dysfunction and prostatic hypertrophy in men. There’s also the brain organ clock cluster of plasma proteins, indicative of a person’s biological brain age versus their chronological age (known as the CognitionBrain aging model), and which was additive to p-Tau-181 for predicting the diagnosis of Alzheimer’s. Reduced diversity of the gut microbiome and presence of certain proinflammatory microbial species have been identified as a risk factor for Alzheimer’s disease. And don’t forget about linked environmental exposures such as air pollution or high consumption of ultraprocessed foods. If all these dimensions of data were integrated, just think how much better we could identify the high-risk group of people years before they manifested mild cognitive impairment, especially in an era in which treatments are being developed to reverse β-amyloid accumulation and reduce brain inflammation, and people are aware of lifestyle characteristics that are associated with reduced risk.
Preventing cancer is another example. Recently, the National Human Genome Research Institute–funded Electronic Medical Records and Genomics (eMERGE) Network published validation of polygenic risk scores with readiness for clinical implementation. Previously, multiancestry, validated polygenic risk scores have been reported for all major types of cancer, including breast, colon, lung, and prostate. In the new report of 10 common chronic diseases, two types of cancer were assessed—breast and prostate—in two large multiancestry cohorts using a threshold of top 5% score for twofold or greater risk of breast cancer and top 10% for twofold or greater risk of prostate cancer. The Mass General Brigham health system began implementing polygenic risk scores for their patient population, with polygenic risk scores for breast, colon, and prostate cancer, identifying with prospective assessment 13% with high risk of breast cancer (more than twofold in white and Black people, not increased among Asian individuals), 6% with high risk of colon cancer (over fourfold in Black people), and 15% with about twofold increased risk of prostate cancer. Polygenic risk scores represent just one layer of data to stratify high risk of a particular cancer. They are inexpensive to obtain from gene chips or low-pass genome sequencing and are complemented by rich genomic data on cancer predisposition genes, such as breast cancer 1 (BRCA1) and BRCA2, which can also pinpoint high risk. It is the combination of common genomic variants that are the basis of polygenic risk scores, and the rare mutations, from whole-genome sequencing, that will provide the most insight into genetic risk.
The electronic health record is rich with information on risk that isn’t currently being tapped. A case in point is the ability to predict pancreatic cancer, notoriously one of the hardest cancers to diagnose in early stages and thus one of the most fatal. Data from the Denmark national registry and the US Veterans Affairs electronic health records were able to differentiate people with 30- to 60-fold increased risk of pancreatic cancer within the next 12 months. Not only was new-onset type 2 diabetes one of the tip-offs, but an AI model that integrated more than 80 features from electronic health records, including lab tests, symptoms, medications, and coexisting conditions, proved helpful for identifying risk of pancreatic cancer. These patterns are generally not discernible to clinicians. Beyond that, there are medical scans, such as a chest computed tomography (CT) scan, which is performed for other reasons, that can reveal pancreatic abnormalities that are frequently missed by radiologists but that AI can more sensitively detect (in one study, 34% sensitivity by radiologists, 93% by AI). Like the example for Alzheimer’s disease, there are many other layers of data to integrate for risk assessment, including the gut microbiome, nutrition, and environmental exposures.
Instead of a multicancer early detection (MCED) blood test (also known as “liquid biopsy”) that only identifies unsuspected cancer in 5 per 1000 people aged 50 years or older, and only 2 per 1000 at early stage (I or II), enrichment for other data that indicate a high risk for cancer should make such tests far more useful. It is the high-risk people who could undergo annual or semi-annual surveillance with MCED for the earliest, microscopic detection of cancer. That would be expected to markedly improve a patient’s prognosis compared with how cancer is screened today, but would have to be proven through randomized clinical trials. [This key point was made this week in a Nature Medicine editorial]
It is important to emphasize that our ability to integrate all these layers of data into multimodal AI models is still at an early stage, and until now has been restricted to electronic health records and genomics, or electronic health records and medical images. High frequency or continuous data from wearable sensors have yet to be incorporated, in addition to many other layers. It’s an analytical challenge that is currently being tackled, and, if and once successful, can be widely applied to other promising forecasting technologies, such as the use of digital twins with nearest-neighbor analysis of vast population-level medical datasets.
Taking lessons from the major progress in AI-powered weather forecasting, which affects our daily lives, there is now an opportunity to greatly sharpen medical forecasting at the individual level to improve long-term health outcomes. All those big, rich data for each individual are being left on the table, unintegrated, not analyzed. Harnessing intensive computing resources and performing large-scale prospective validation studies can get us to where we should be for identifying people at high risk for major diseases, arming them with ways to preempt or mitigate their occurrence.
Here are multiple new reports from this week on different steps towards accurate medical forecasting:
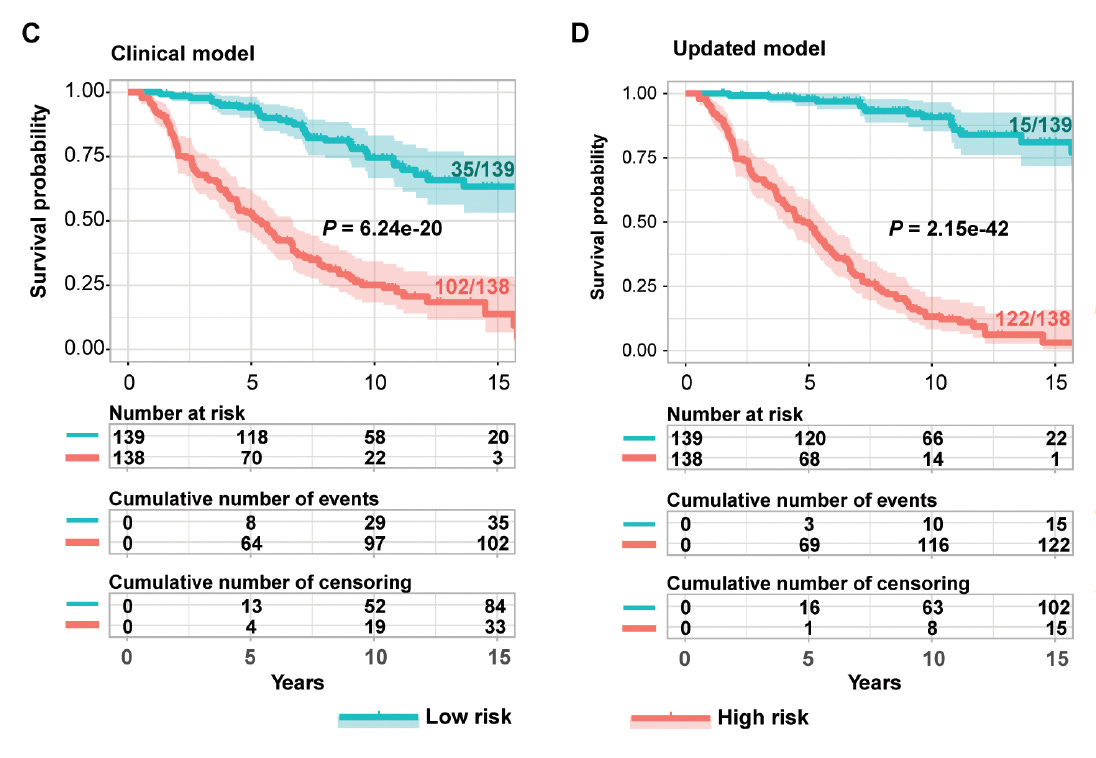
Multiple omics (DNA methylations, proteins, micro-RNAs, gene variants) for predicting kidney disease and failure in people with Type 1 diabetes compared with the standard clinical model for risk
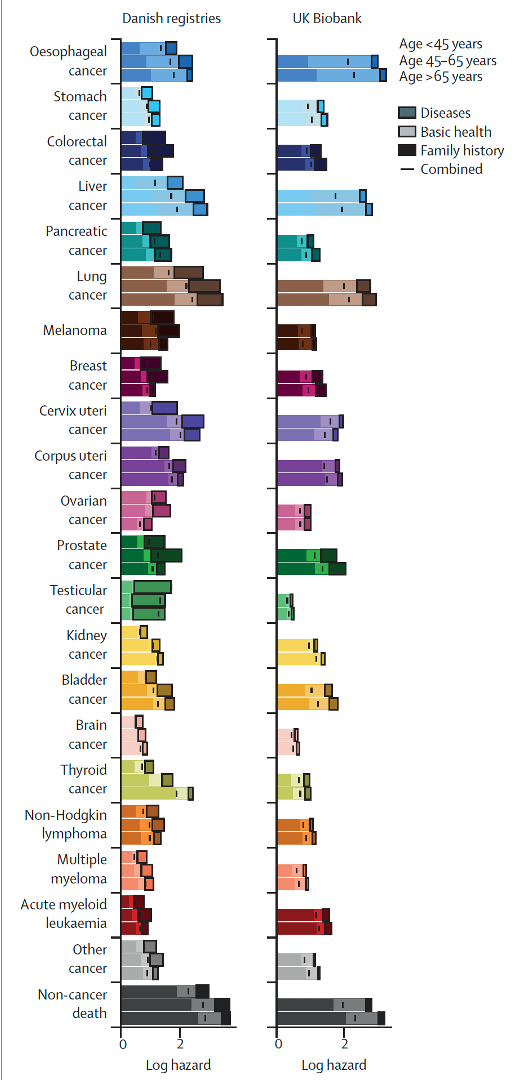
Predicting multi-cancer risk over 3-years using a national health resource (Denmark)
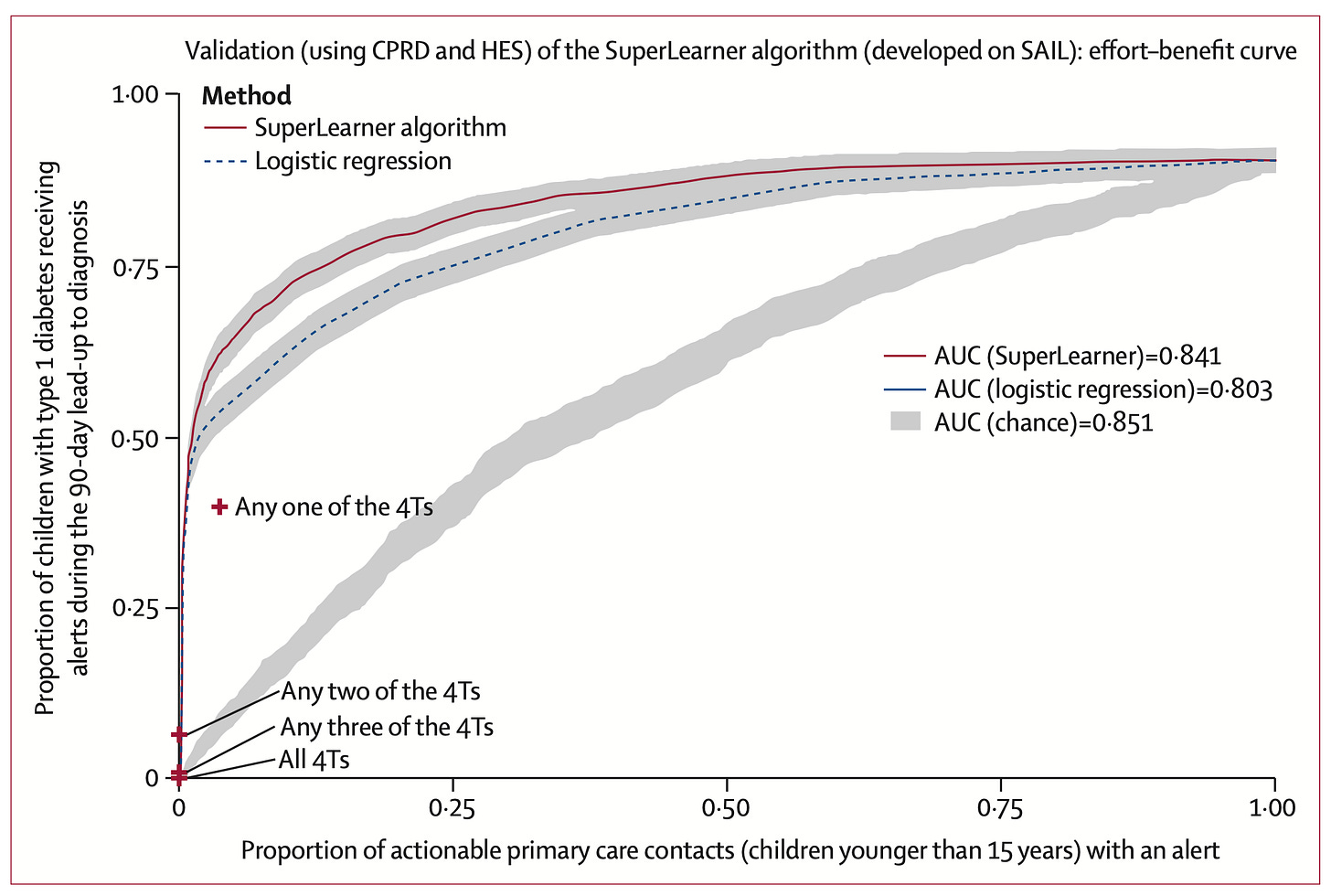
3. Predicting Type 1 diabetes and timing in children
Summing Up
Identifying high-risk individuals for major medical conditions —which have evidence for actionability for prevention— is in the works. There are a variety of ways to improve accuracy of forecasts incorporating multiple layers of a person’s data, fully integrating the data via multimodal A.I.. A goal is not just to partition a high-risk status for a specific disease, but also the likely timing the condition may manifest, since, for example, just carrying “high-risk” but its likely occurence after age 90+ has much less significance. Similarly, the added knowledge of likely timing applies as to when to initiate an intervention.
Historically, in medical practice, we have not done well at primary prevention or detection of cancer at the earliest, microscopic state to enhance the success of therapy. Instead, we are reactively geared towards secondary prevention after a heart attack, stroke, atrial fibrillation, or diabetes have occurred, or picking up cancer at late stages. Efforts to push us towards highly accurate medical forecasting have the chance to change this pattern. Like everything else it will require validation, not just providing models, but that it works to change the natural history of diseases—achieving primary prevention of cardiovascular metabolic, neurodegenerative diseases or very early detection of cancer.
*********************************
The Ground Truths newsletters and podcasts are all free, open-access, without ads.
Please share this post if you found it worthwhile.
Voluntary paid subscriptions all go to support Scripps Research. Many thanks for that—they greatly helped fund our summer internship programs for 2023 and 2024.
Note: you can select preferences to receive emails about newsletters, podcasts, or all I don’t want to bother you with an email for content that you’re not interested in.


Fascinating. Longitudinal data in the non-universal healthcare US not so great; VA probably best bet. Huge potential; I actually thought (stupid I know) electronic medical records would help form the basis for improved disease understanding but their purpose is billing. Available to life insurance companies? Health insurance companies? Employers? There's the rub.
So helpful. Powerful, too -- to see the NOA use case and understand how possibke it is today to develop the large computational models you highlight. One difference between the weather and model and what is needed in healthcare: public access to large data sets. Despite federal law to ensure patient access to personal data, the friction for accessing large healthcare data via silo'd systems is daunting. Might this suggest a different G2M for US "mass computational" efforts, or a need to go to large exUS systems to prove the model (India). Interesting to consider merist of ex-US system, which may offer broader data sets for correlation (UBI, etc.)?