Multimodal AI for medicine, simplified
The catalytic impact of large language models (Generative AI)
AI in medicine is basically a single mode story to date. Help reading an X-ray or MRI, finding polyps during a colonoscopy, provide patient coaching for a specific condition like diabetes, or a preliminary diagnosis of a skin lesion or heart rhythm from a smartwatch recording. It has largely been image-centric to date, with minimal integration with, or use of inputs from, text and voice. But over time that narrowness and constraint may well be alleviated.
My colleagues and I recently wrote a review of the potential for multimodal AI, when data from many sources can be ingested and processed as seen below. No one has yet done this yet: pulling together and extracting the knowledge from individuals at scale—sources of data that include multiple continuous biosensors, biologic layers such as the genome and microbiome, environment, and medical records. That ultimately will not only be attainable but will enable many opportunities such as the virtual health coach, hospital-at-home, and a digital twin infrastructure.
How do we get there?
There has been unparalleled interest in ChatGPT and large language models (LLMs). It’s been referred to as an iPhone moment by many, with the most rapid user base ever seen, comparators to 100 million users below.
I see it as major 4 building blocks that have brought us to here. Deep learning AI was potentiated by a large number of graphic processing units (GPUS or TPUs), but generative AI is that on high dose steroids. This has created seemingly insatiable ability to ingest data, in the form of tokens, with parameters, the term representing the number of connections between neurons, and the computing power metric known as floating point operations (FLOPS). Rather than supervised learning which largely powered medical algorithms to date, requiring expert annotation of images and ground truths, the training with LLMs is disproportionately limited relative to the massive data inputs. The new GPT-4 model details were released today It’s multimodal (now bimodal with text and image integration) capability has increased: “outperforming existing LLMs on a collection of natural language processing tasks”
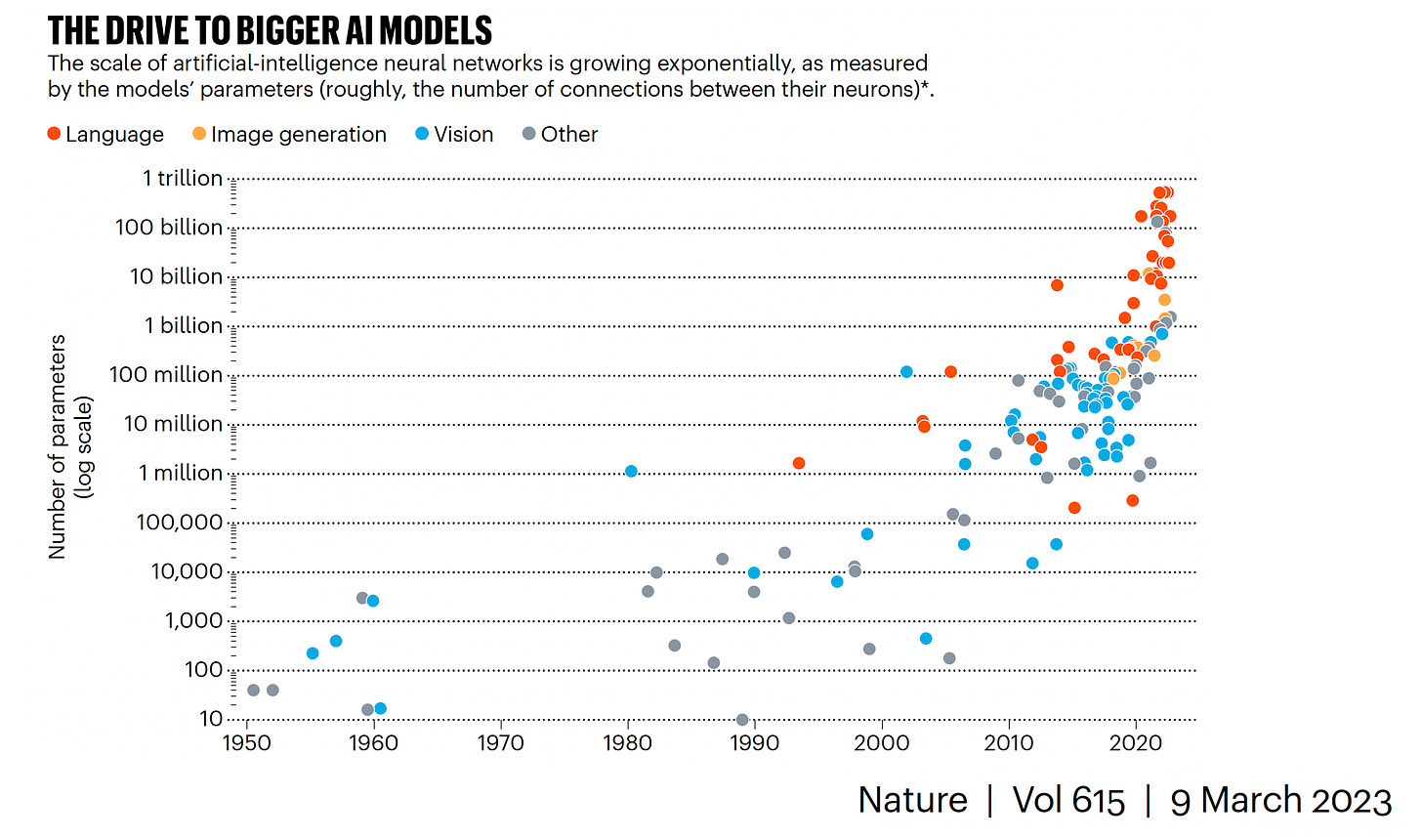
It’s the coming together of multimodal data this quite an advance for LLMs, as you can see the acceleration of each modality increasing separately on a log-scale below
Is Bigger AI (More Parameters) Better?
Graph below from Anil Ananthaswamy’s recent and excellent Nature piece showing the accelerated evolution (log-scale) for each data domain (by parameters).
The answer to that question is clearly no. While transformer models, as used with LLMs, have already surpassed 1 trillion parameters as seen by Xavier Amatriain’s graph below
the number of tokens is exceptionally important, as seen in the X-axis below.
Perhaps the most successful LLM for transformative impact to date has been DeepMind’s AlphaFold which accurately predicts 3-D structure of proteins from amino acid sequences. So the rumor that GPT-4 would have >100 trillion parameters is not only wrong, but markedly over-values one of the components of LLMs. Saying it again: tokens are damn important. Bigger (by parameters) isn’t necessarily better.
From Chatting with Sydney to Multimodal Medical AI
While the lengthy conversation that Kevin Roose had with Sydney (Bing’s new search integrating ChatGPT+) will go down in history and even made it to the front page of the New York Times, remember that we’re still in the early days of LLMs and virtually none have had specific or extensive pre-training in medicine.
I recently wrote about how Google’s Med-PALM and ChatGPT did well on the US medical licensing examination. Today we’ve just learned that the next iteration of Google’s LLM, Med-PaLM-2 scored an 85%, well above the previous report of 67% (60% is passing score threshold). But that’s using a chatbot’s memory for a single mode language task that chiefly relies on memory and the statistically driven juxtaposition of words, aptly described as a lossy JPEG image by Ted Chiang in the New Yorker (a piece not to miss).
There’s a big jump forward from this to drive keyboard liberation for clinicians, which is just now beginning, using LLM training inputs from millions, or tens of millions, of medical records. In the months ahead we’ll see the beginning of generative AI to take on so many language-based tasks: synthetic office notes based on voice (with automated prescriptions, next appointments, billing codes, scheduling of labs and tests), pre-authorization from insurance companies, aggregating and summarizing a patient’s history from scouring their medical record(s), operation and procedure notes, discharge summaries, and more. Examples from Doximity (docsGPT) and Abridge are showing us the way. As opposed to the electronic health record disaster that has transformed clinicians to data clerks and led to profound disenchantment, over time LLMs may well be embraced as the antidote.
Obviously there are major issues that have to be grappled with, that not only include LLM hallucinations (mandating humans-in-the-loop oversight), pseudo-reasoning, amplification of concerns over bias, privacy and security of data, the dominance of the tech industry and more. But, without LLMs, it would be hard to see how we could get multimodal AI forward progress. It’s ultimately the ability to move seamlessly between medical images, text, voice, and all data sources (sensors, genome, microbiome, the medical literature) that will afford the many opportunities shown in the top diagram of this post.
For that, I’m excited to see the rapid evolution of LLMs and their future application for medicine and healthcare.
Thanks for reading and sharing Ground Truths!
The (Support) link below, opening this newsletter to paid subscribers, is explained in my previous post