Spatial biology is lighting it up
A rapid fire chain of discoveries are illuminating health and disease
This week was the culmination of a series of biologic discovery mapping papers that are rapidly changing the way we understand health and disease. In the current issue of Nature, and various Nature journals (including Nature Cell Biology, Communications Biology, Nature Communications, Scientific Data), along with Science, there was a capping off a substantial number of reports in recent years, providing new insights about the ~37 trillion cells in the human body and how they interact at the cellular and molecular levels. It’s a complex topic that I’ve held back on writing about for some time, but it’s clear that spatiotemporal biology has become a foundation for future biomedical advances. Everyone with an interest in science should know about it!
There’s been intense interest and commitment for defining the biology at the cellular level antedating the new reports, such as the Tabula Sapiens project which reported in May 2022 on single-cell and single-nuclei RNA sequencing for over 1 million cells, covering 500 cell types, across more than 30 human tissues.
The field of spatial biology is becoming a critical driver of life science discovery. For decades we knew that cancer tissue was heterogeneous, with some cells exhibiting more aggressive growth and spread features than others, but we didn't have the tools to understand it. Now we do, and it’s well beyond understanding the spatiotemporal evolution of cancer—it’s being applied across the board from Alzheimer’s disease, atherosclerotic plaque, the retina, pancreatic B-islets, spinal cord to whole organ characterization such as heart, kidney, lung, intestine, and maternal-fetal interface. Much of this work is part of the Human BioMolecular Atlas Program (HuBMAP), now in its production phase involving 60 institutions, over 400 investigators, to generate high-resolution 3D accessible maps for > 20 organs. You can think of it as generating the Google Map for the entire healthy adult human body at the single-cell level. And it’s quickly becoming a reference, anchor resource for diseased cells and tissue.
How Does it Work?
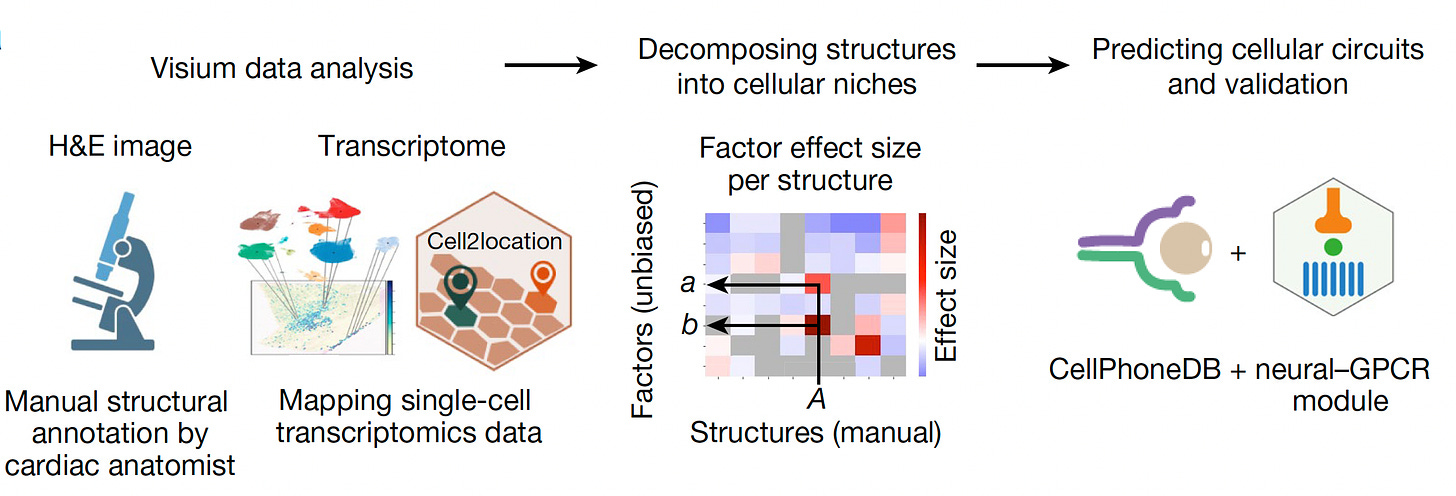
Here’s a rudimentary graphic for what it entails. A slice of tissue of interest is put on a specially prepared microscope slide (top, left), different omics are defined (RNA/transcriptomics, DNA, proteomics, epigenomics, such as chromatin) which are essentially barcoded and can be spatially analyzed (right panel, below). The matched cells to the image, in this tumor example, are 2D reconstructed to provide space and time relationship. (Yes, it’s more complicated than this with micro-dissection, laser capture tools, but it’s the same principle.)
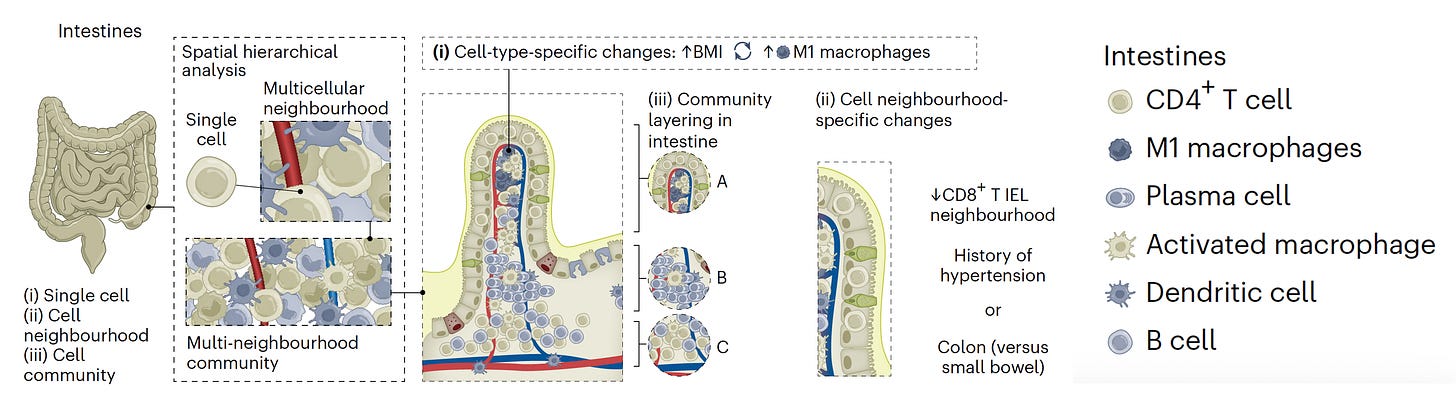
As illustrated in the new HuBMAP for the intestine, this analysis yields a multi-level representation of the various cell types (including rare and previously not known), along with the hierarchical organization and context of the neighborhoods, communities, and functional tissue units. Not shown here is that this architectural digest can go subcellular for high-resolution of intracellular organelles (such as mitochondria).
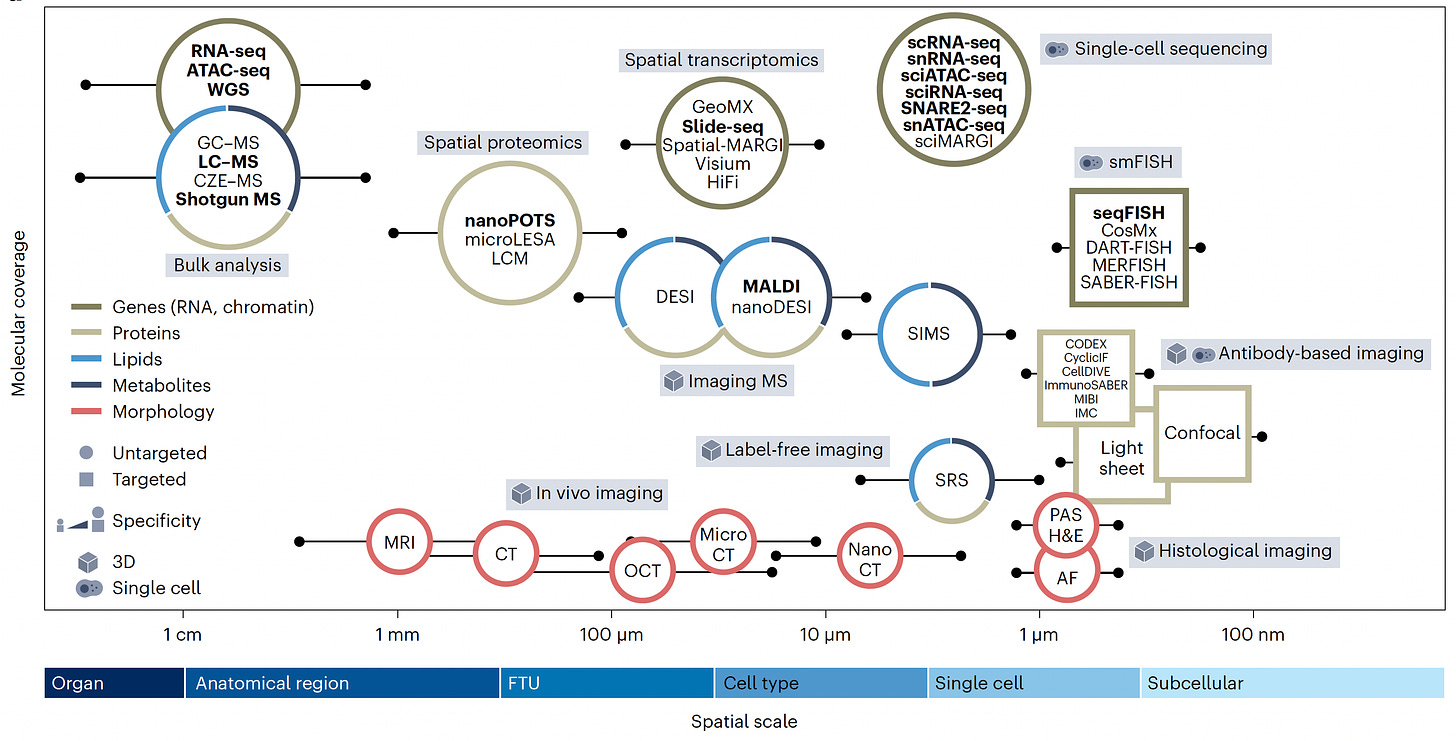
This is seen in the graph below (from Jain et al) with the many types of single cell or single nuclei sequencing that can be performed across the spatial scale (x-axis), the multiplex mapping of the cell’s in situ constituents. Intact, label-free (leveraging A.I.) and in vivo imaging can be obtained through different techniques, along with histology or antibody-based imaging that complements the omic sequencing for deep characterization. Talk about scale: in several studies of cancer spatiotemporal evolution, more than 1.1 to >1.6 million cells were mapped by at least some of these methods!
OK, so what?
Here are some recent (and I find fascinating) discoveries for medical conditions and in healthy tissues that were made with spatial biology
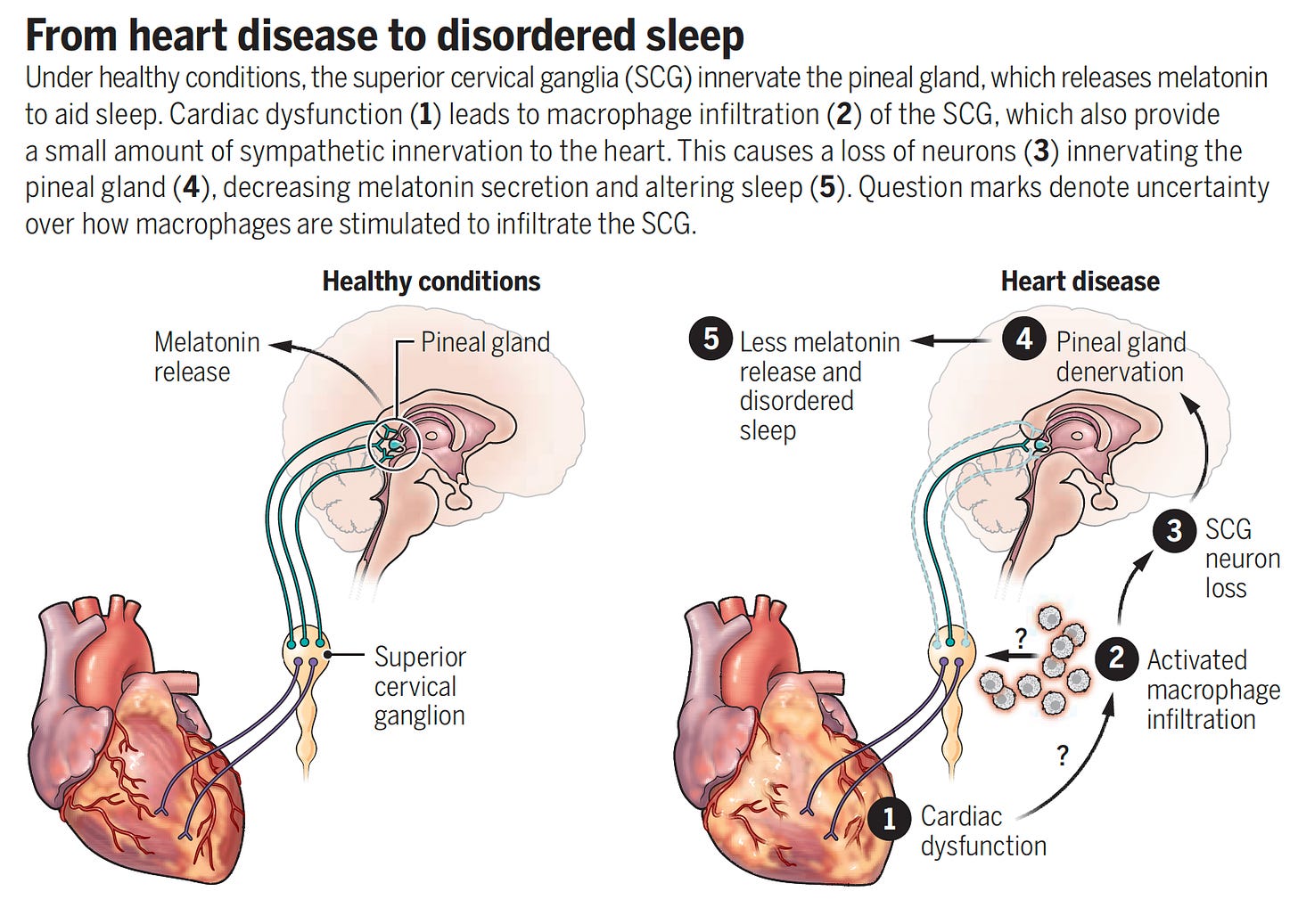
—The superior cervical ganglia (SCG) and pineal gland loss of innervation (immune-mediated) leads to sleep disturbance in people with heart failure
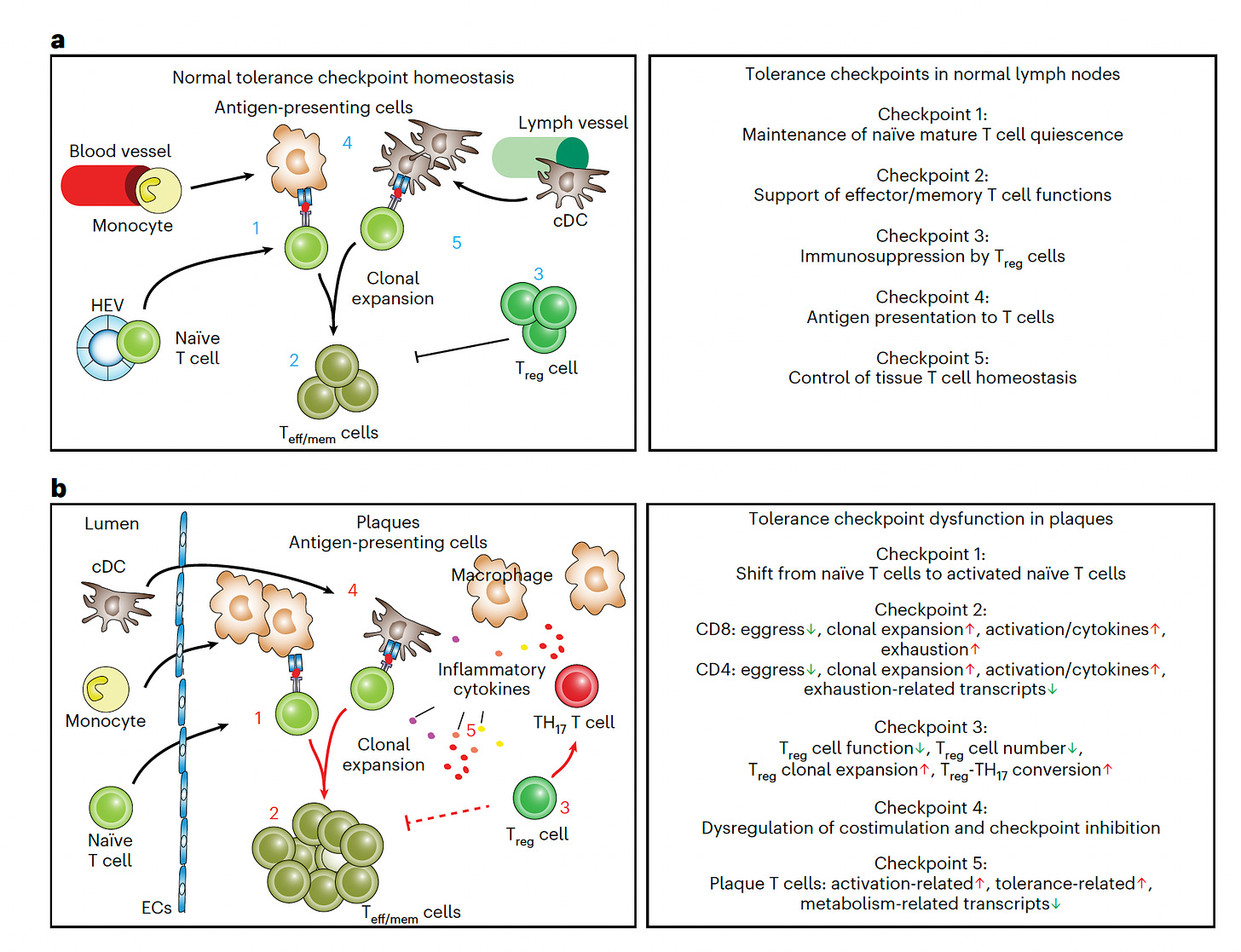
—Atherosclerotic plaques have important T-cell features of autoimmunity
—The brain’s immune system (the skull bone marrow-meninges axis niche reservoir) and “enormous amounts” of the SARS-CoV-2 spike protein localized to the skull bone marrow, meninges and brain tissue in people who had Covid but died of other reasons, fully discussed here
—The heart has glial cells (weren’t they supposed to be only in the central nervous system?) that interact with the pacemaker, has cells that express the target of GLP-1 (the anti obesity drugs). And the epicardium has plasma cells forming an immunological niche
—That the intestine in people with hypertension have reduced CD8+ T cells
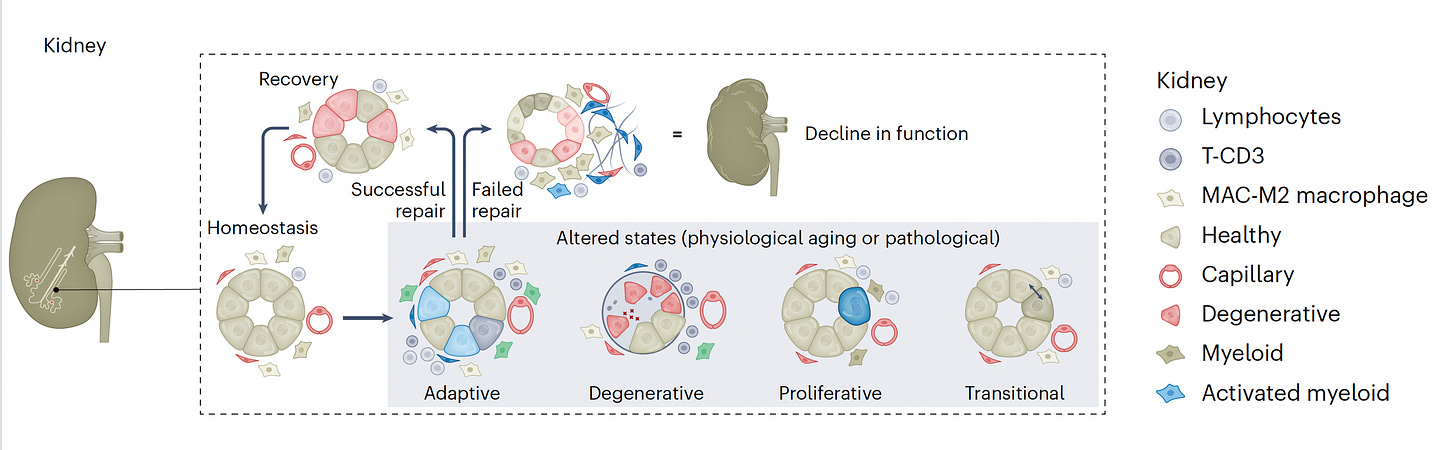
—Features of cells in the end-stage heart failure that recover or what is inducing scarring (fibrosis) after kidney injury, including specifically by kidney stones
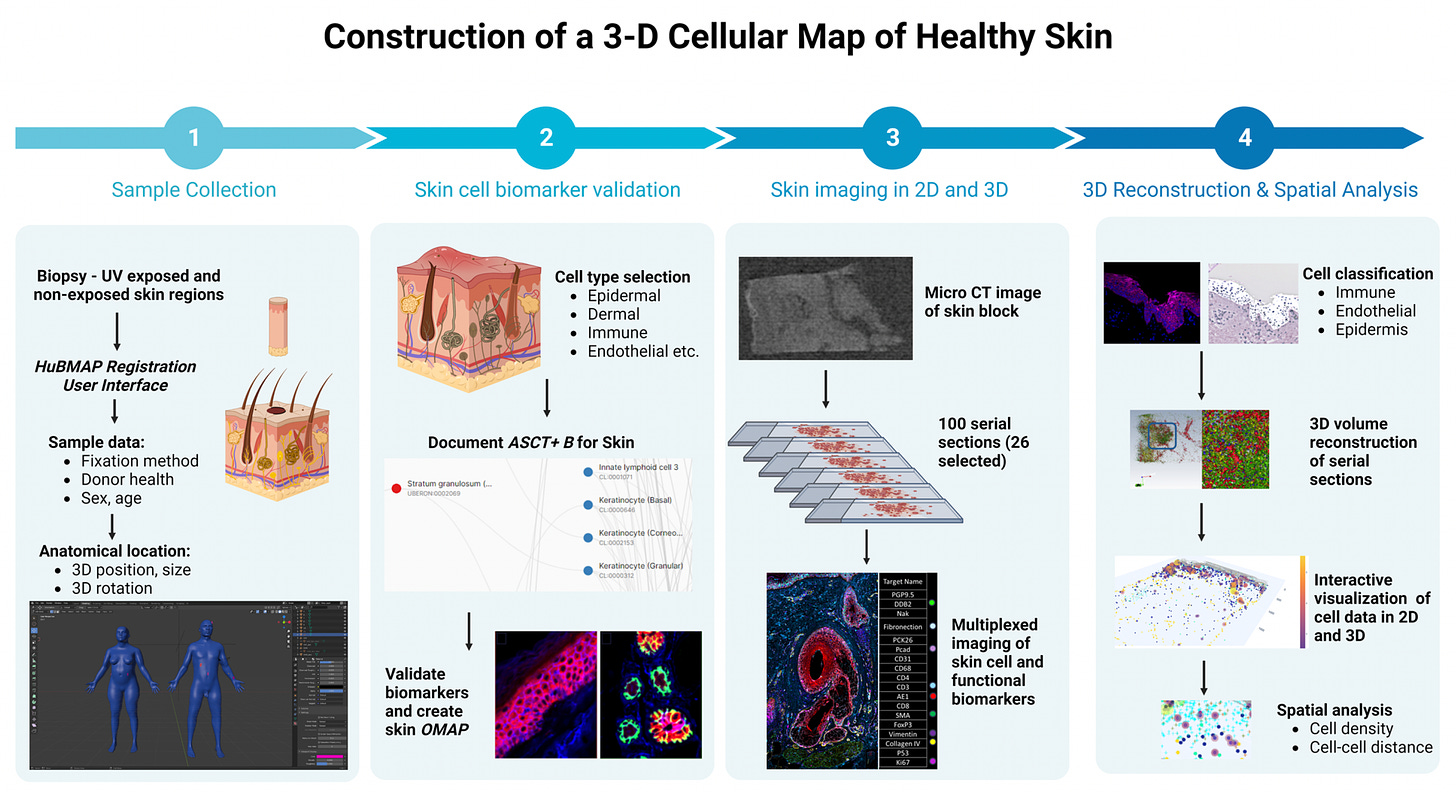
—How skin cells are damaged by sun exposure in 3D
—How pancreatic β-islet cells are changing in the evolution of Type 2 diabetes
—Unraveling the different cells, genes, pathways and dysfunctional processes underlying Alzheimer’s disease
Summary
From the examples above (there are plenty more), I hope I’ve convinced you that spatial biology is helping us make extraordinary advances in biomedicine. The studies are a way to establish cause and effect relationships that are often quite difficult to come by, to trace evolution (or development) of a tissue, organ, or cancer, and to come up with surprising findings on the functional importance of rare cells or neighborhoods of cells. What used to be called the microenvironment, clones and sub-clones within tissue can all now be precisely characterized at high resolution single-cell level. No more “heterogeneity”—define it! Biomarkers, such as a distinct immune response like tumor infiltrating lymphocytes, can be derived from maps that track evolution, no less for potential discovery of patterns that align with therapeutic outcomes.
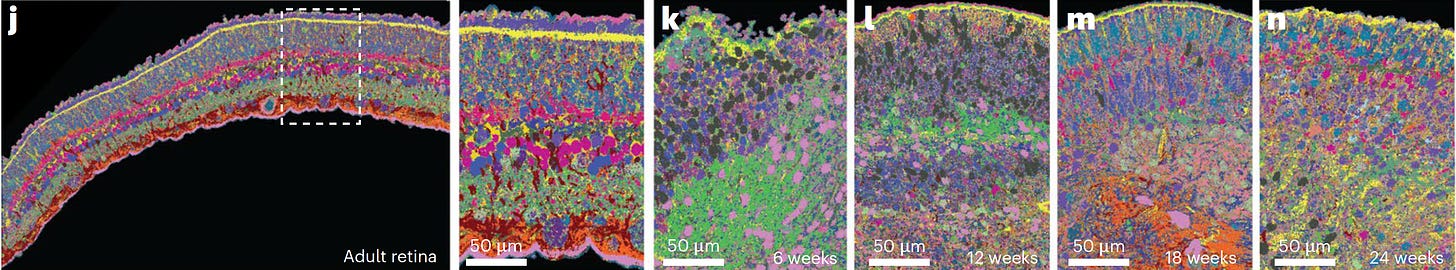
I haven’t even yet mentioned the aesthetic aspect—the beauty of these maps—such as the development of the human retina being spatiotemporally traced below.
With multi-level omics and imaging the datasets are typically immense (think multiplexing a million cells) and machine learning/deep learning tools with automation are necessary for analytics. That’s yet another major contribution of A.I. to life science.
But remember we’re still in the early stages of this work. The HuBMAP has an ambitious workplan over the next 3 years with several new technologies, get more sub-cellular, more 3D, and develop better A.I. and visualization tools as summarized in the Figure below.
You’ll be hearing a lot more about spatial biology in coming years. I hope this brief, simplified review is a primer for the outpouring of more exciting and fascinating advances in life science and medicine that lie ahead.
Thanks for reading Ground Truths!
Your free subscription denotes your support of this work. Should you decide to become a paid subscriber you should know that all proceeds go to support Scripps Research. That has already helped to bring on several of our summer high school and college interns.
\