Cancer Science in On a Tear
New insights on evolution, resistance, and therapies
The past 2 weeks of the journal Nature were packed with 16 new publications that have substantially advanced our understanding of cancer biology. In this issue of Ground Truths, I’ll review some of the salient points and themes from this extraordinary crop of research findings.

Mapping Cancer in 4 Dimensions With Spatial Omics
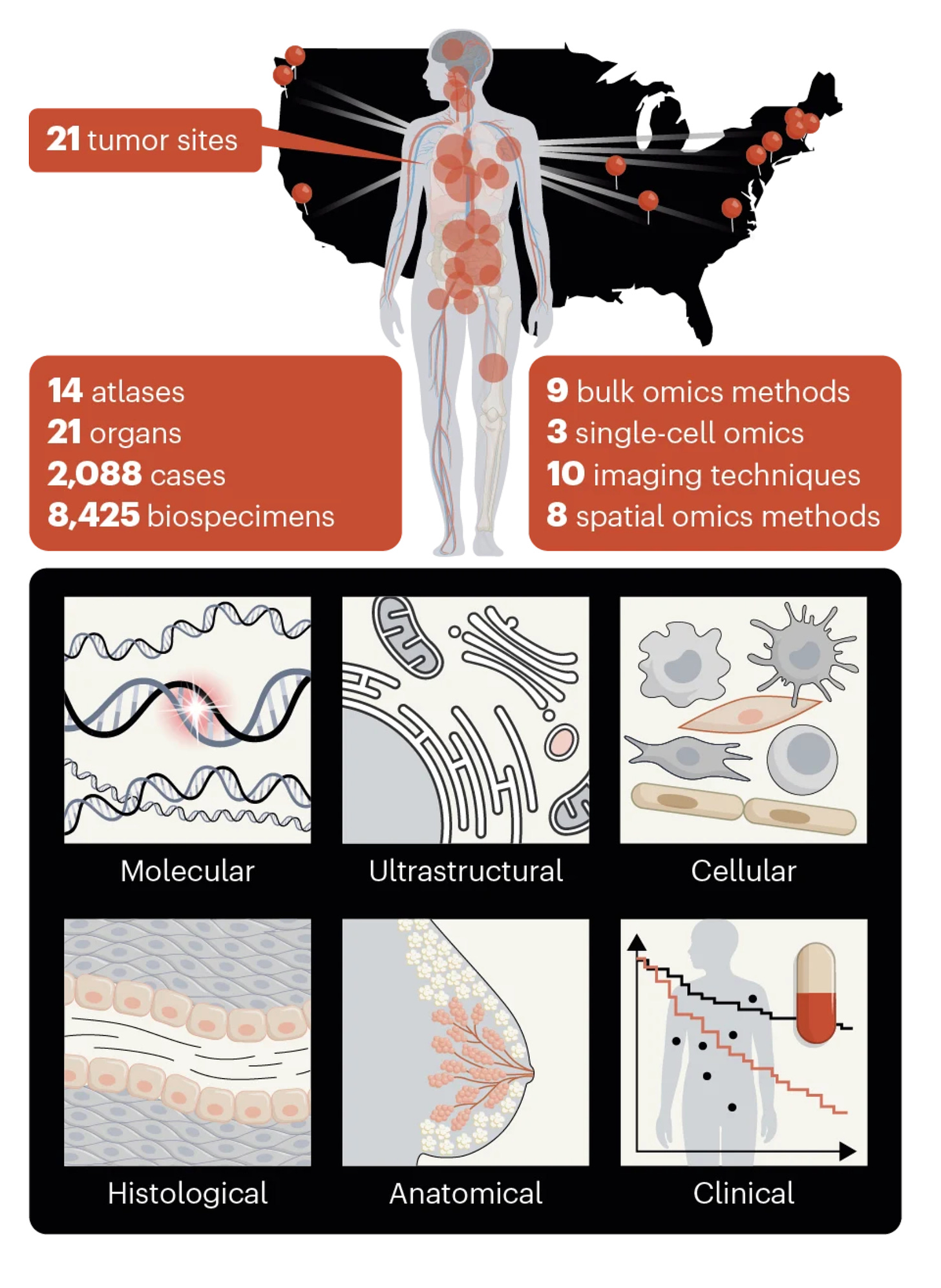
The Human Tumor Atlas Network (HTAN) is a National Cancer Institute funded initiative involving 10 research centers that began its work in 2018. Its overall mission has been to improve our understanding of how cancer originates, progresses, resists or responds to therapies via high resolution mapping of cellular and molecular processes. In particular, spatial omics (a topic I’ve reviewed here and here) was a theme in multiple papers, highlighted by the cover above. It provides mapping by space and time (spatiotemporal) making it 4-D and by doing so yields new insights about tumor genesis and spread.
A collection of the new 12 papers on ~20 types of cancer, from more than 2,000 individuals, at multiple levels (as shown above) represents a formidable body of work, to say the least. The journals included not just Nature but also Nature Cancer, Nature Medicine, Nature Methods, and Communications Biology.
I won’t attempt to go through all of these papers here, but try to hit the highlights.
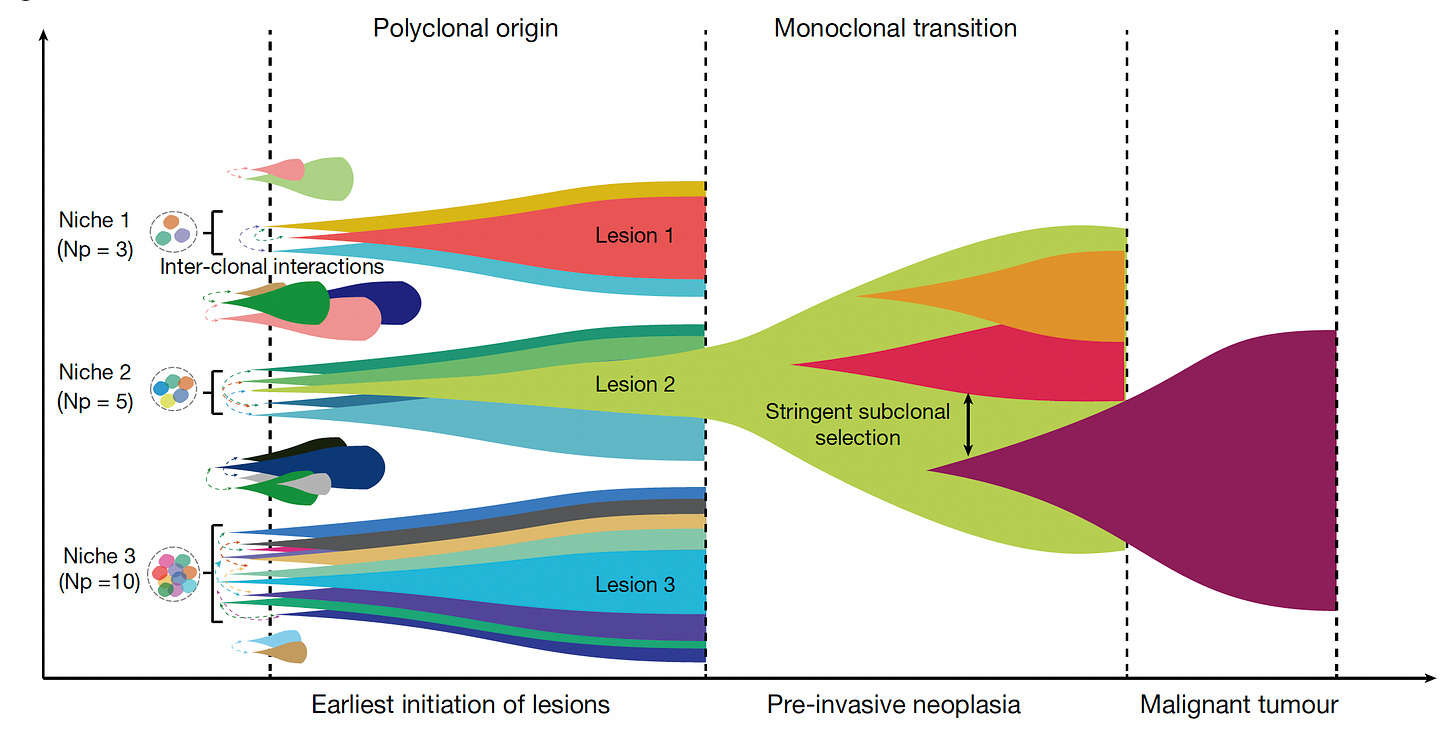
For the origin of colon cancer, the use of single-cell sequencing with base editor CRISPR barcoding led to the finding that there are multiple clones of cells (up to 10, polyclonal) that cooperate and winnow down to one at the pre-cancerous stage. This work was complemented by another single cell CRISPR platform—enabling a molecular clock—also showing multiple clones in 15-30% of colonic pre-cancers.
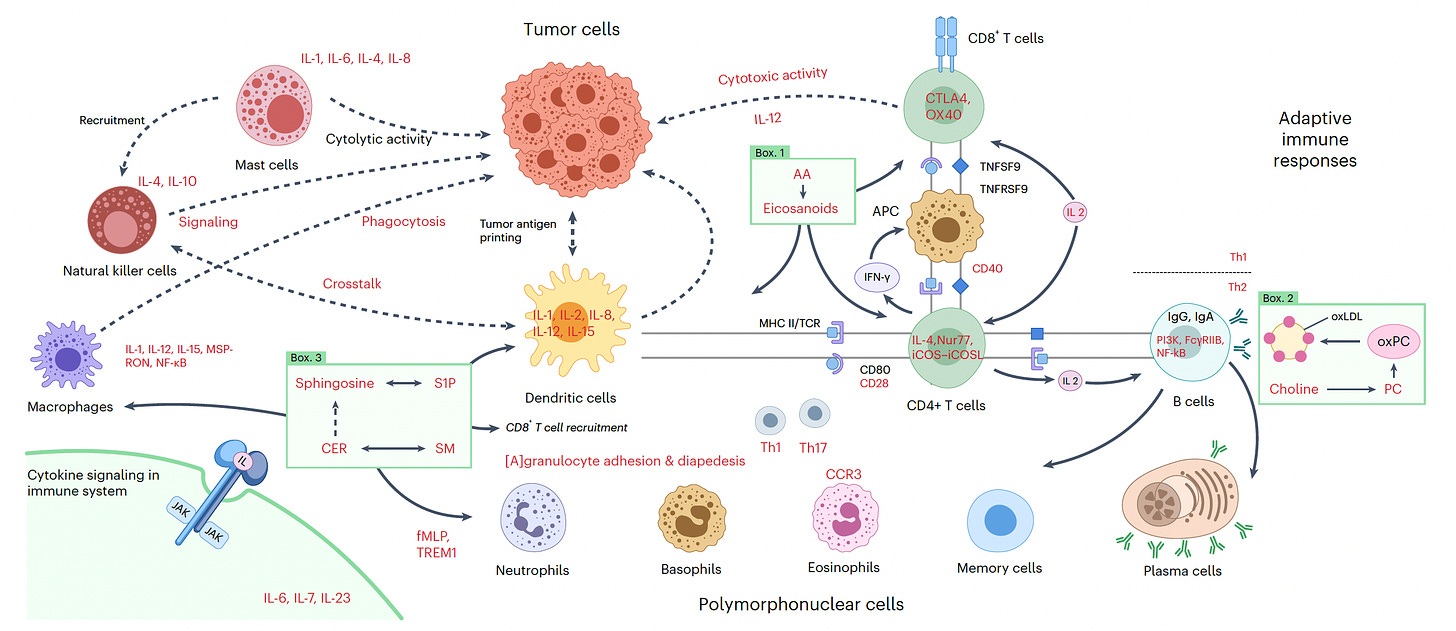
Also on colon cancer was a remarkably high resolution of familial adenomatous polyposis, a heritable form of colon cancer. The layers of omics data integrated include the genome, transcriptome, proteome, metabolome, methylome, and lipidome. This work also traced the activated immune response from the transition from mucosa to dysplastic polyp, as graphically represented below.
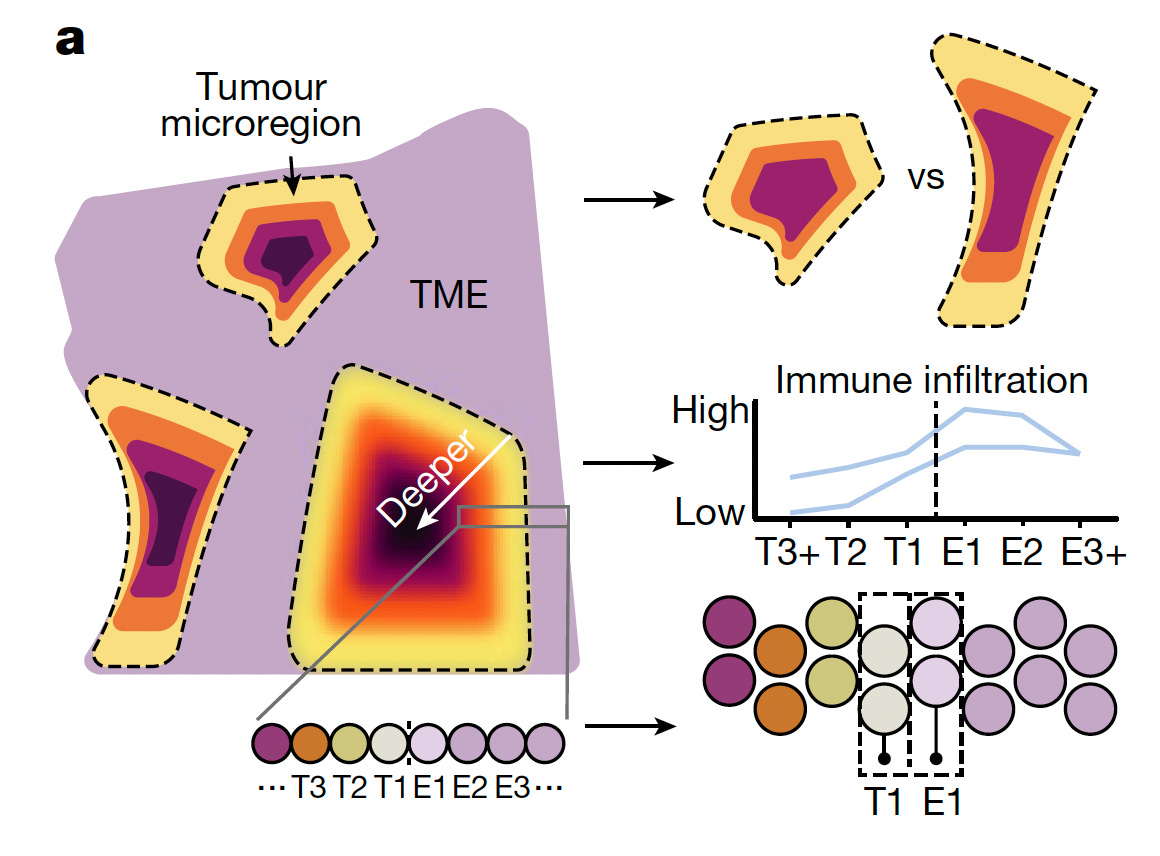
For mapping breast cancer and insights about how the immune response looks, a helpful short video is worth a review. Below the tumor microenvironment (TME) from is seen with variable immune cell infiltration, which, when low, suggests T cell exhaustion and inability to mount an adequate response (what was called “hot” and “cold” neighborhoods”) That could theoretically be countered with various types of immunotherapy. However, in parallel, another report of 60 women with aggressive breast cancer illuminate the dynamic immune response that materially changed when assessed through serial biopsies.
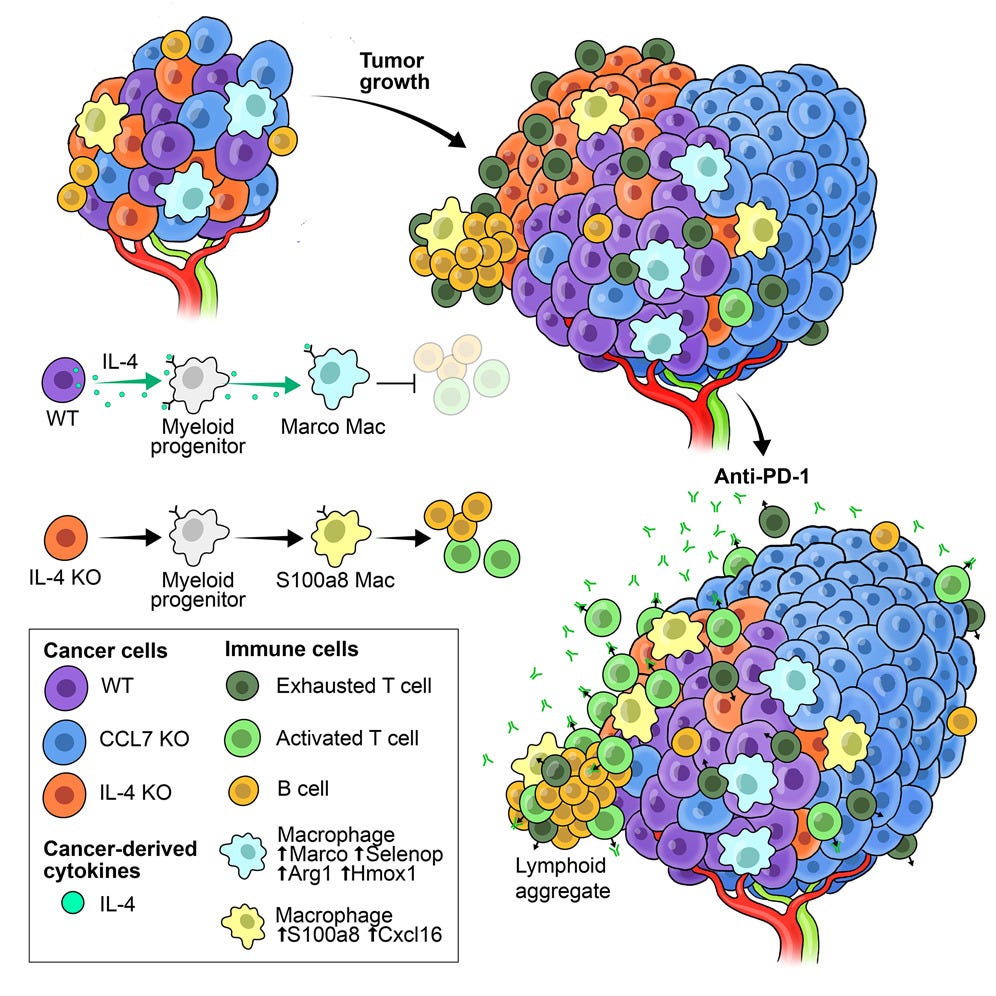
That same week, apart from the bltiz of HTAN papers, there was one in Cell on ovarian cancer. Using spatial omics (again with CRISPR screens, known as Perturb-map) it probed why ovarian cancer is so resistant to immunotherapy. It uncovered cancer cell derived interleukin-4 (IL-4) as a driver of this resistance to immune checkpoint inhibitors (i.e. anti-PD-1). This raises the opportunity to target IL-4 to overcome immune suppression.
Extrachromosomal DNA (ecDNA) and Cancer
The other batch of 4 papers in this week’s Nature were dedicated to ecDNA. The cover (Top Cover Figure, Outside Influence) gives you a sense of what these look like—circular DNA particles that are found in the nuclei of cancer cells that are particularly pernicious, bad actors. They contain both gene and regulatory regions (such as promoters, enhancers) that can interact with each other, and rapidly rev up tumor progression. While they can occur early in the evolution a cancer, their presence was more than 7-fold higher in Stage 4, 2.8-fold higher in Stage 3 than in Stage 1 cancers. Their association with poor clinical outcomes is linked to both promoting the growth/spread of tumor and blocking the immune response, a double whammy if there ever was one.
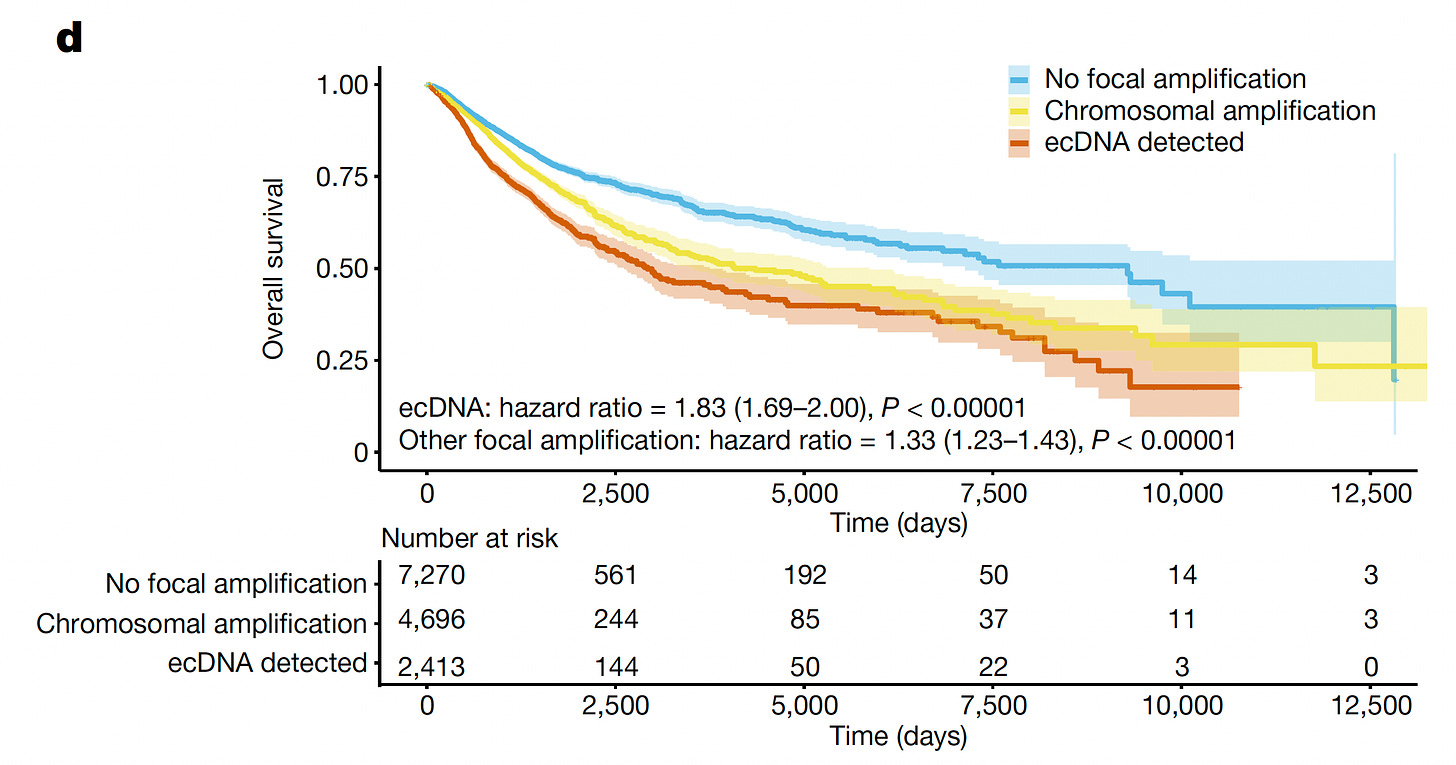
The most comprehensive body map of ec-DNA in cancer was derived from about ~15,000 patients via 13 UK NHS Genome Medicine Centers, an outgrowth of the Genomics England 100,000 Genomes Project. 17% of cancers had ecDNA which carried a poor prognosis for survival (graph below). Notably, the ecDNA containing tumors were more likely to be seen after cytotoxic chemotherapy. Their content varied by tumor type, with the most regulatory DNA in colorectal and most oncogene (cancer-driver gene) in brain tumors. Of note, HER2+ breast cancer had an unanticipated high proportion of ecDNA (in ~39-54% of these cancers).
Other findings on extrachromosmal DNA honed in on the “jackpot effect” of ecDNA coordination to achieve their deleterious impact, their role in bladder cancer arising in response to chemotherapy and their propensity to form structural variants.
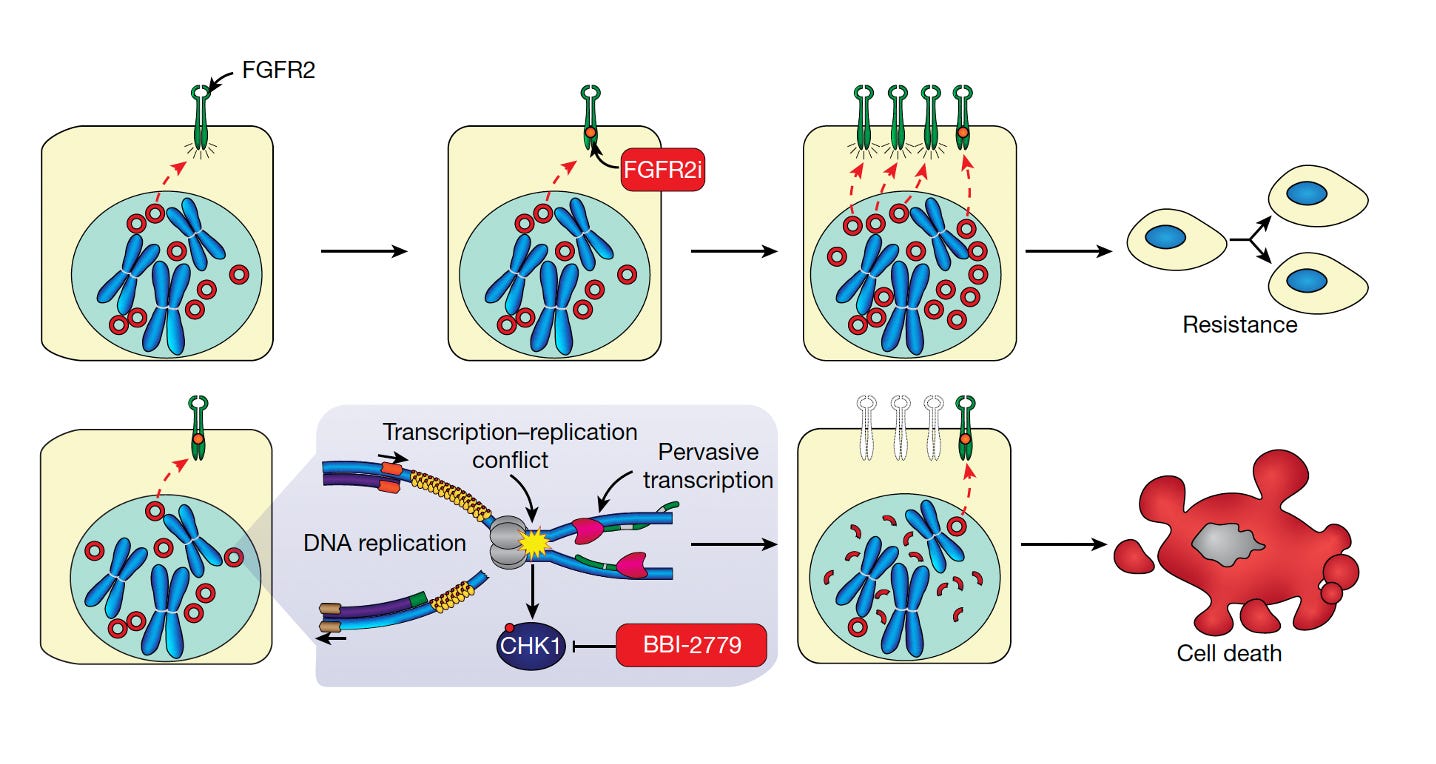
Perhaps the most notable paper in this cluster raised the potential for a specific ec-DNA therapy. The Stanford group led by Howard Chang presented data for a potent, highly selective oral inhibitor of ec-DNA (BBI-2779) that selectively kills cancer cells that contain these circular DNAs. While very early, this conceptually may turn out to represent a breakthrough via promoting cancer cell self-destruction. Its through enhancing their transcription-replication conflict, schematically shown below with the CHK1 inhibitor. I particularly liked their powerful summary statement: “Our work demonstrates the feasibility of a synthetic lethality of cancer-specific cellular excess to turn the molecular advantages of ecDNA in cancer against itself.”
High-Resolution Cancer
In aggregate, these reports are showing the way towards understanding cancer at a much higher level of resolution than ever before. The new multi-dimensionality is not just through the lens of time and space, not just cancer cells, immune cells, and the tumor microenvironment, not just integration of all the omics (DNA, RNA, proteins, metabolites, lipids, chromatin, methylation). It’s all of these at once!
The dedicated, global, and coordinated efforts are making major inroads to unravel the complexity of cancer. I can’t remember any point previously when this many new publications in leading journals came out this rapidly, ushering in new ideas for the origin and progression of cancer, no less potential therapies.
This is all part of a bigger movement towards high-resolution humans, that has been enabled by the same tools—single-cell and single-nuclei sequencing, multi-omics, CRISPR barcoding for molecular clocks and perturbations, and A.I. analytics. It’s reflected by my recent post on the dawn of spatial medicine where deep visual proteomics at the single cell level led to a lifesaving treatment.
I hope you will see this as I do, an especially exciting and unique time in life science and medicine, with convergence of many tools and analytics that have been incubating, now making possible an acceleration of information and knowledge. Our unprecedented capability to define human biology at the pan-molecular, cellular, and tissue levels, and see things that were previously not discernible, is laying the foundation for important progress.
******************************************************
Thanks for reading and subscribing to Ground Truths.
If you found this interesting please share it!
That makes the work involved in putting these together especially worthwhile.
All content on Ground Truths—its newsletters, analyses, and podcasts, are free, open-access.
Paid subscriptions are voluntary. All proceeds from them go to support Scripps Research. Many thanks to those who have contributed—they have greatly helped fund our summer internship programs for the past two years.
Added note: Many of you have abandoned the X platform for reasons that I understand. While I intend to continue to post there because of its reach to the biomedical community, I will post anything material here in the Notes section of Ground Truths on a daily basis.