Antedating the world’s notice of large language models (aka generative A.I.) with ChatGPT in November 2022 was AlphaFold2, published in the previous year. I wrote about it extensively in a prior GT edition. A quick summary: it was the 2021 Science Breakthrough of the Year, solving the >50-year protein folding problem. With the capability of predicting 3-D structure for >200 million proteins from amino acid sequence, made open-source, now having been used by more than 2 million researchers in 190 countries, AlphaFold2 quickly transformed life science research. It was also recognized by the 2024 Nobel Prize in Chemistry to Demis Hassabis and John Jumper, along with David Baker for A.I. protein design work.This has clearly been an A.I. achievement that stands out as one of—if not the—most impressive one in science to date.
But it turns out it was also a notable precursor and template for many large language models in life science that has now moved to a hyper-accelerated phase. In fact, at least 10 have been published in the past couple of weeks! The goal of this edition of Ground Truths is to review the progress we’re making with a pace of A.I. advances in life science that I’ve not seen before, or frankly even imagined would be possible.
Below are the covers of the leading journals in science over the past couple of weeks. The Evo model was published in Science in the 15 November issue and the Human Cell Atlas Foundation model was published in Nature on the 21 November issue. If you’ve been reading Ground Truths over the past few years, you know I pay attention to science covers because so much work often goes into them to visually capture a major milestone or breakthrough.
AI for Science Forum
In London this week, Google Deep Mind and the Royal Society of Medicine organized the AI for Science Forum conference. The conference had been planned months previously, but the timing, in the midst of this unprecedented surge of new A.I. life science publications, was especially apropos.
Here is the video of the panel that I moderated on “Science in the Age of AI” with Pushmeet Kohli, who heads up A.I. for Science at Google Deep Mind, Alison Noble, Professor at Oxford University, and Fiona Marshall, who directs Novartis Research. If you have a chance to review it, you’ll get the balance expressed between excitement and concerns for how fast things are moving.
At the conference, there was an inspiring intro video (only 2.5 minutes) and an especially memorable conversation between Hannah Fry, a mathematician and phenomenal moderator, and Demis Hassabis. That is followed in the same video by a fun panel with John Jumper, Demis, Jennifer Doudna, and Sir Paul Nurse (4 Nobel laureates).
AlphaFold 3: Predicting Proteins Plus
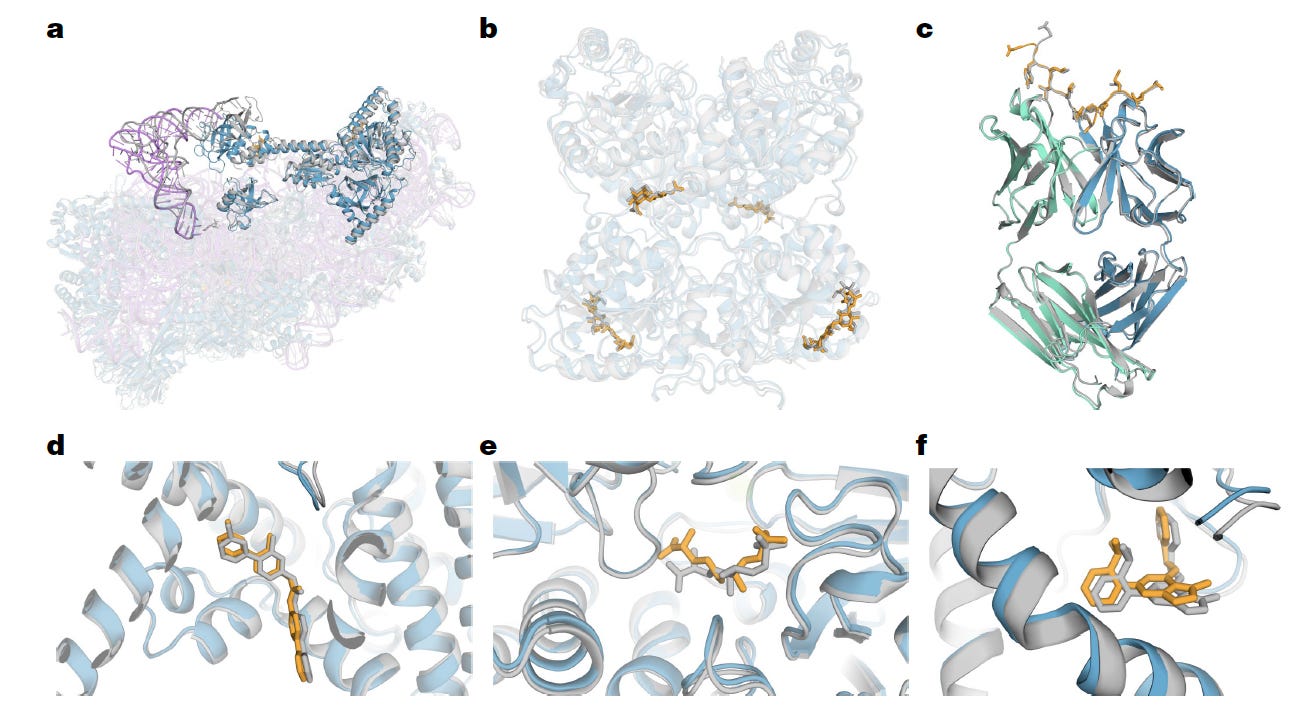
In May 2024, AlphaFold 3 was published, going well beyond predicting protein structures. The upgraded model (now diffusion-based) accurately predicts the 3D structure of proteins in complex with other proteins, DNA, RNA, small molecules and ligands (some examples seen below) directly from sequences. Notably, its prediction for 80% of protein-ligand complexes was within 2 Angstroms (1 Angstrom is 0.1 nanometer, or 1 ten-billionth of a meter) experimental error. This represents a major step forward for understanding how biomolecules interact with one another.
Initially, it was not open-source, the code was withheld, as were the model weights, stirring a controversy because that went against the journal (Nature) policy. But on 11 November 2024, a week before the conference, AlphaFold 3 was made available to the research community, but not for commercial use.
New Large Language of Life Models (LLLMs)
Let me briefly review the new models here. You’ll note I’ve added an L to the usual LLM to denote it pertains to life science.
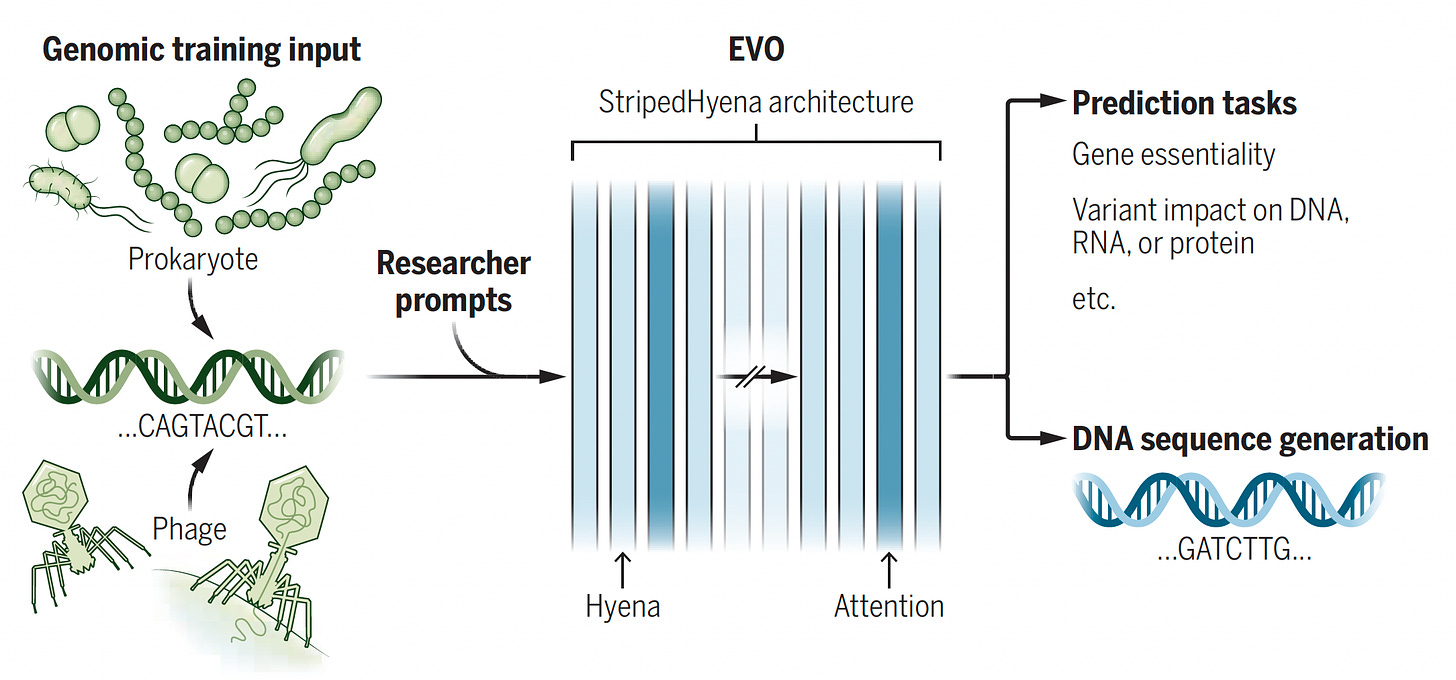
Evo. This model was trained with 2.7 million evolutionary diverse organisms (prokaryotes—without a nucleus, and bacteriophages) representing 300 billion nucleotides to serve as a foundation model (with 7 billion parameters) for DNA language, predicting function of DNA, essentiality of a gene, impact of variants, and DNA sequence or function, and CRISPR-Cas prediction. It’s multimodal, cutting across protein-RNA and protein-DNA design.
Figure below from accompanying perspective by Christina Theodoris.
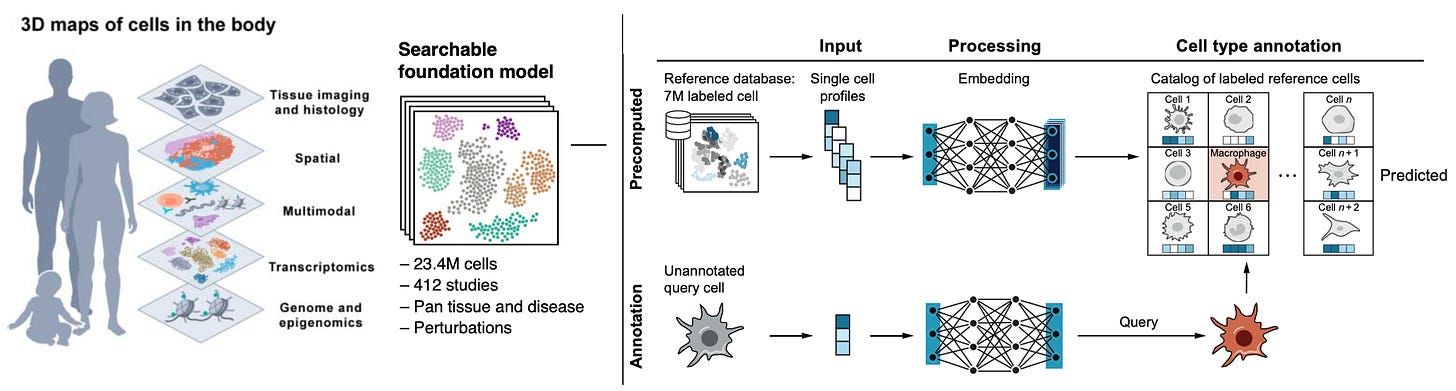
Human Cell Atlas A collection of publications from this herculean effort involving 3,000 scientists, 1,700 institutions, and 100 countries, mapping 62 million cells (on the way to 1 billion), with 20 new papers that can be found here. We have about 37 trillion cells in our body and until fairly recently it was thought there were about 200 cell types. That was way off—-now we know there are over 5,000.
One of the foundation models built is Single-Cell (SC) SCimilarity, which acts as a nearest neighbor analysis for identifying a cell type, and includes perturbation markers for cells (Figure below). Other foundation models used in this initiative are scGPT, GeneFormeR, SC Foundation, and universal cell embedding. Collectively, this effort has been called th “Periodic Table of Cells” or a Wikipedia for cells and is fully open-source. Among so many new publications, a couple of notable outputs from the blitz of new reports include the finding of cancer-like (aneuploid) changes in 3% of normal breast tissue, representing clones of rare cells and metaplasia of gut tissue in people with inflammatory bowel disease.
BOLTZ-1 This is a fully open-source model akin to AlphaFold 3, with similar state-of-the-art performance, for democratizing protein-molecular interactions as summarized above (for AlphaFold 3). Unlike AlphaFold 3 which is only available to the research community, this foundation model is open to all. It also has some tweaks incorporated beyond AlphaFold 3, as noted in the preprint.
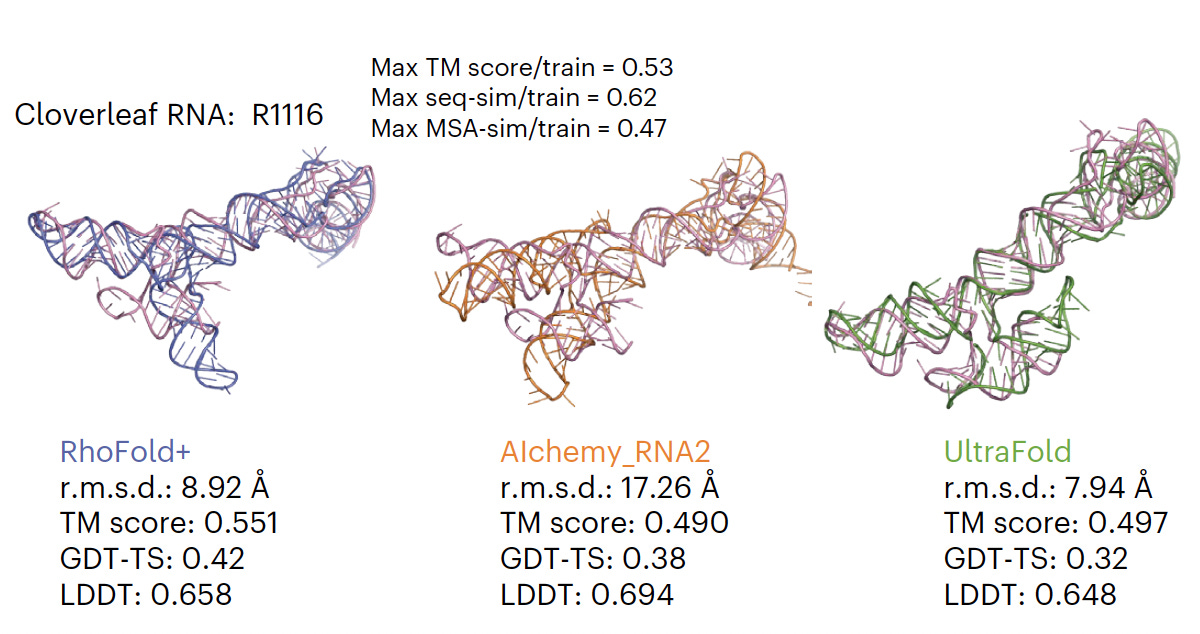
RhoFold For accurate 3D RNA structure prediction, pre-trained on almost 24 million RNA sequences, superior to all existing models (as shown below for one example).
EVOLVEPro A large language protein model combined with a regression model for genome editing, antibody binding and many more applications for evolving proteins, all representing a jump forward for the field of A.I. guided protein engineering.
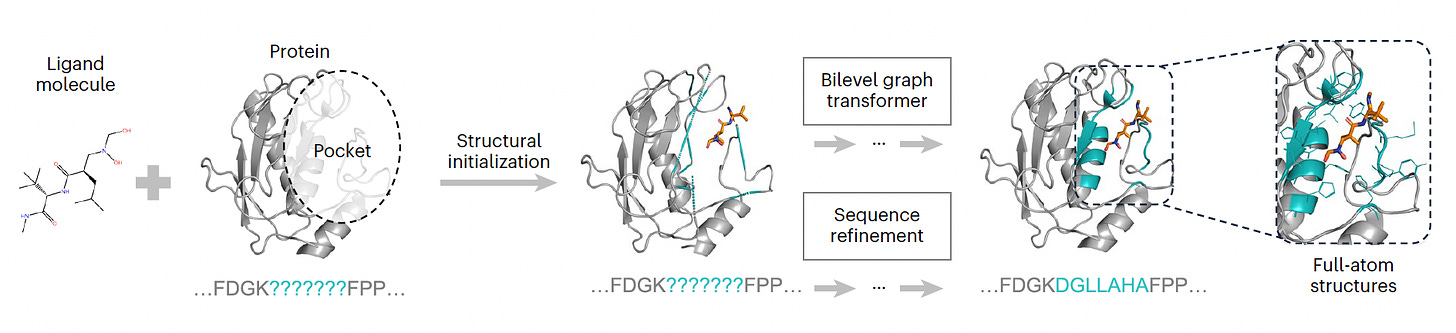
PocketGen A model dedicated to defining the atomic structure of protein regions for their ligand interactions, surpassing all previous models for this purpose.
MassiveFold A version of AlphaFold that does predictions in parallel, enabling a marked reduction of computing time from several months to hours
RhoDesign From the same team that produced RhoFold, but this model is for efficient design of RNA aptamers that can be used for diagnostics or as a drug therapy.
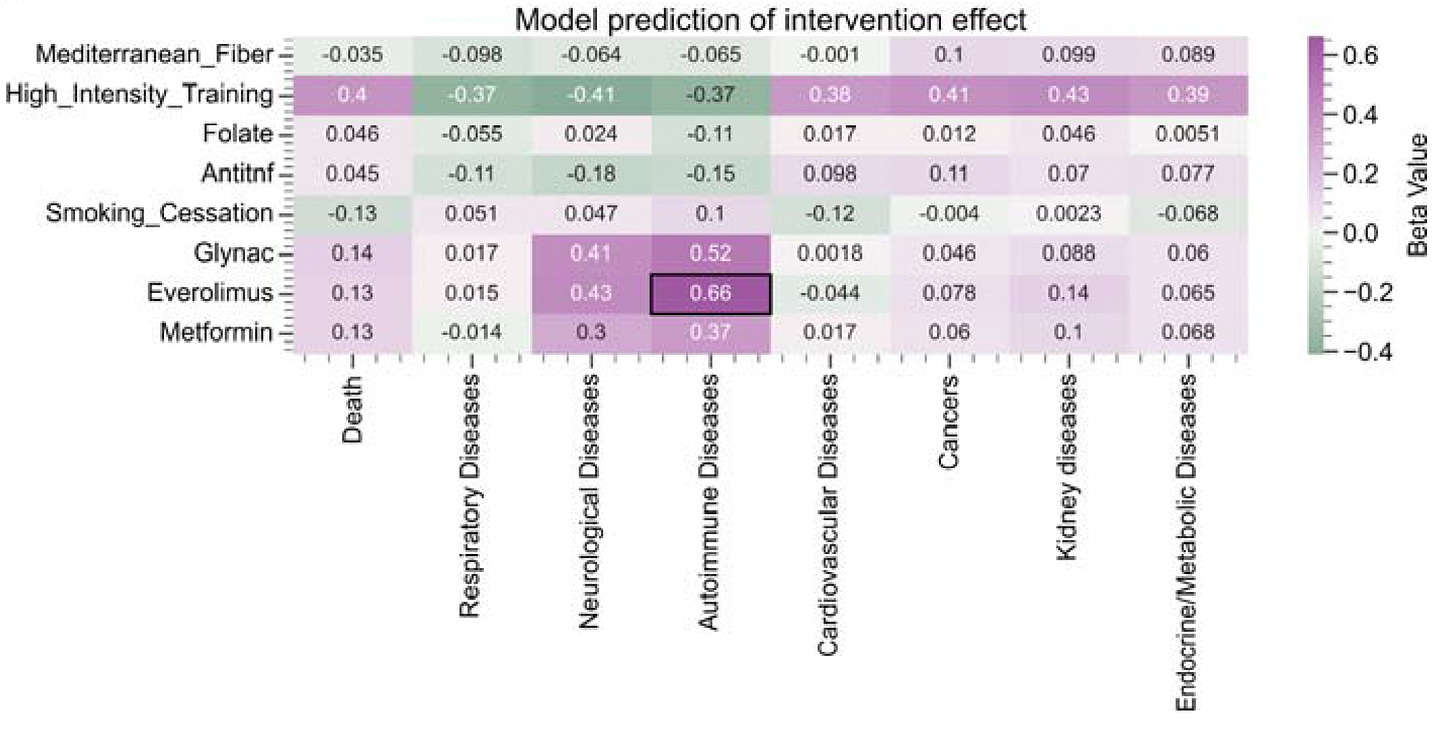
MethylGPT Built upon scGPT architecture, trained on over 225,000 samples, it captures and can reconstruct almost 50,000 relevant methylation CpG sites which help in predicting diseases and gauging the impact of interventions (see graphic below).
CpGPT Trained on more than 100,000 samples, it is the optimal model to date fo predicting biological (epigenetic) age, imputing missing data, and understanding biology of methylation patterns.
PIONEER A deep learning pipeline dedicated to the protein-protein interactome, identifying almost 600 protein-protein interactions (PPIs) from 11,000 exome sequencing across 33 types of cancer, leading to the capability of prediction which PPIs are associated with survival. (This was published 24 October, the only one not in November on the list!)
The Virtual Lab
This one really grabbed me, since we are just learning about the power of deploying an autonomous agent to get tasks done. James Zou and colleagues at Stanford took it to another level, charging a group of 5 agents, each with domain-specific backgrounds (as seen below), to work together on a project designing nanobodies directed against the SARS-CoV-2 virus. With frequent independent lab meetings of the 5 agents, and minimal human supervision, the LLLM multi-agent team came up with 2 potent new nanobodies, potentially representing a new pathway to discovery in science.
Synthesis
What we are witnessing here is extraordinary, to say the least. As a close follower of the progress in life science over the past few decades, this work, in aggregate, surely qualifies for the label of unprecedented, occurring at a breakneck pace. The breakthrough of AlphaFold2 and the transformer architecture of generative A.I. has clearly inspired scientists all over the world to apply LLMs to all components of life, be it proteins, DNA, RNA, ligands, not to just understand structure and biology at the atomic level, but also evolution and design. Unlike medicine, there is no regulatory bottleneck for using these models (as discussed in the video above), most of which have been democratized, made open-source. That will only further catalyze the field of digital biology, which recently led to the discovery of Bridge RNAs (see my recent podcast with Patrick Hsu, “A Trailblazer in Digital Biology”).
As Demis Hassabis pointed out in London at the AI for Science Forum, the implications here go well beyond life science, as we’re seeing “engineering science—you have to build the artifact of interest first, then use the scientific method to reduce it down and understand its components.” Much work still has to be done to fully understand these complex neural networks, with deep interrogation, like an fMRI of the brain.
Are we now entering an exponential phase of digital biology, a vertical takeoff, a golden age of discovery? Time will tell, but think about what happens when you have a sum of the parts. When you have exquisite knowledge of the structure of proteins and all their biomolecular interactions (other proteins, DNA, RNA, ligands), not just pairwise but in multiples, in pathways, but (not just static) also with perturbations, along with all the big epistemic gains being made at the cellular level. This sets up the potential for the virtual cell in the future to simulate and predict its function. Perhaps that work will ultimately be undertaken, at least in part, by a virtual lab!
I hope you can see the potential this body of work has for advancing our understanding of human biology and physiology, health and disease processes, and what is already taking off— designing and developing new therapies. It’s exhilarating!
*******************************
Thank you for reading and subscribing to Ground Truths. As I’ve learned recently talking with subscribers, many people still don’t realize that it’s a hybrid of newsletter/analyses like this, and podcasts, roughly alternating between each format.
The Ground Truths newsletters and podcasts are all free, open-access, without ads.
Please share this post with your friends and network if you found it informative!
Voluntary paid subscriptions all go to support Scripps Research. Many thanks for that—they have greatly help fund our summer internship and educational programs.





Wow- between the innovations you've highlighted here and the the emerging concept of the holobiont, we'll have a completely different and better understanding of the biologic systems we all live within.