The under-appreciation of CHIP
It's time to drop the "indeterminate" and get serious about this biomarker
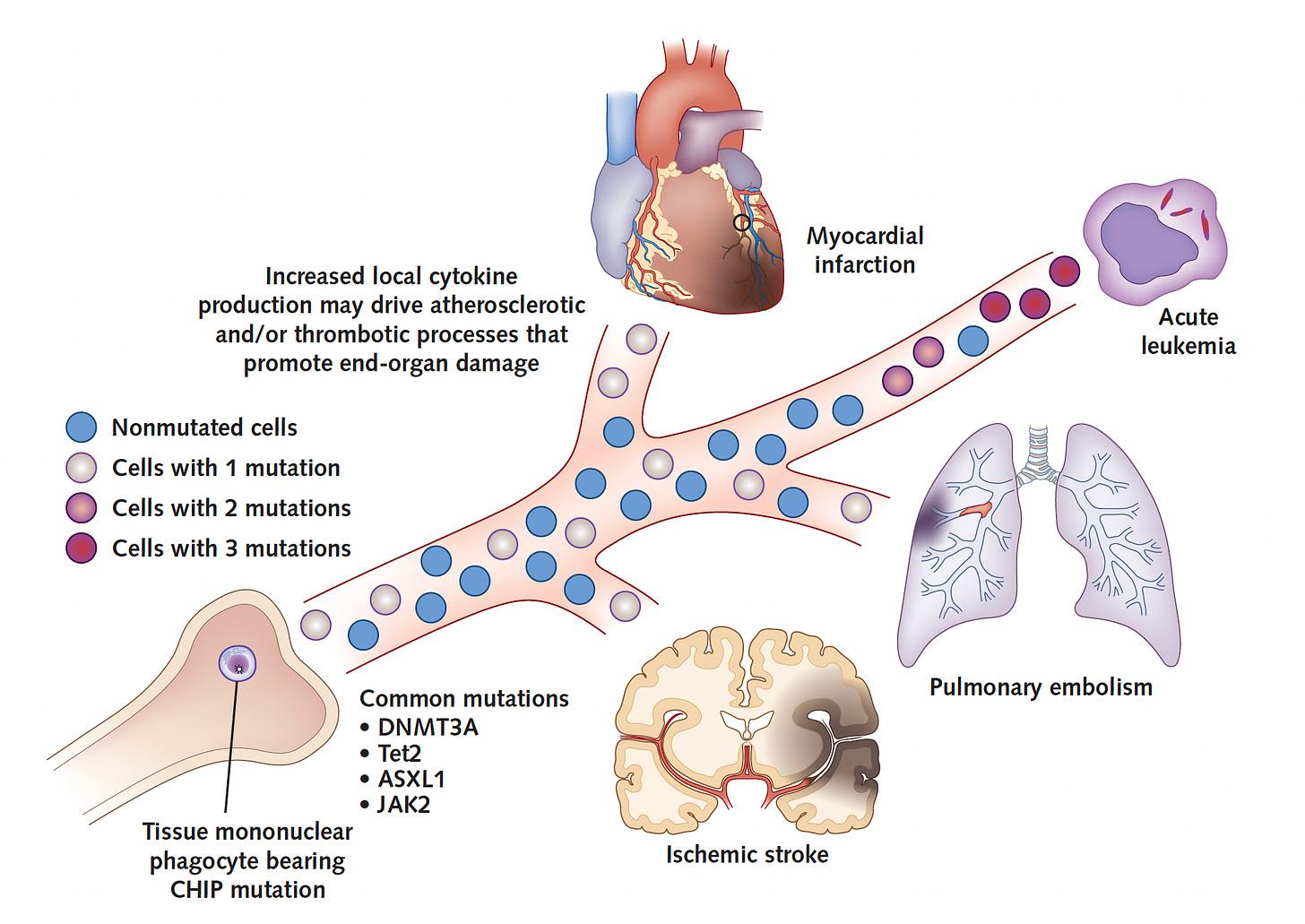
Each day the 50,000 to 200,000 blood stem cells in our bone marrow, known as hematopoietic (HSC), produce billions of specialized blood cells—red blood cells, platelets, B and T lymphocytes, and various myeloid cells (white blood cells, such as neutrophils and monocytes). As we age, the repeated cell divisions of the HSCs lead to acquired (“somatic”) mutations. When such a mutation of an HSC leads to a clone that expands—which denotes some fitness advantage— it is known as clonal hematopoiesis. CHIP, which stands for clonal hematopoiesis of indeterminate potential, refers to presence of myeloid blood cells with a driver gene mutation (with a frequency of ≥ 2%) without any clinical criteria of a blood cancer. It turns out CHIP is a major biomarker, not just for blood cancer (an 11-fold risk of a blood malignancy, or absolute 0.5% risk per year), but also for heart disease, blood clot events such as stroke and pulmonary embolism, and many other chronic illnesses (simplified Figure below).
We’ve known about age-related CHIP for more than a decade, and over that time it has become clear that it is not only common, but has expanded associations with multiple chronic illnesses, especially the whole spectrum of cardiovascular conditions. With the conventional threshold of 2% of blood cells carrying a specific mutation(s), over 10% of people age 70 and older have CHIP and that proportion goes up substantially with more advanced age. A schematic of a specific CHIP mutation in the TET2 gene, the 2nd most common driver mutation seen in CHIP, is shown below
The Figure below for CHIP prevalence is based on whole genome sequencing from over 97,000 people. We know that people of Hispanic or East Asian ancestry have about half the incidence of CHIP, adjusted for age, than other ancestries, and that smoking is associated with an 18% increase rate of CHIP. The larger the CHIP clone, the greater the risk.
But the prevalence of CHIP depends of what sequencing method is used—WGS=whole genome, WES=exome, or targeted to a specific gene, ECS-error corrected sequencing) and what variant allele frequency (VAF) is selected. Irrespective of the threshold and method used, you can see CHIP, in the Figure below, becomes increasingly pervasive over the lifespan.
It turns out 75% of the driver genes in CHIP are DNMT3A, TET2, and ASXL1, as shown below. (To avoid getting into too much complexity and into the weeds, I won’t go into detail about these genes that are involved with DNA methylation and epigenetic modifications, but this link provides the details if you are interested.) There is a specific relationship between the driver gene and risk of clinical conditions. Furthermore, the appearance of certain driver mutations like DNMT3A can be traced from multiple decades earlier, whereas others have much faster evolution and are associated with worse clinical outcomes. It’s interesting that some clones can sit stagnant for many years, and then take off, whereas others show steady growth. While 90% of people with CHIP have only 1 driver mutation, the other 10% with more than one mutation are at increased risk of adverse sequelae, including acute myeloid leukemia and heart disease.
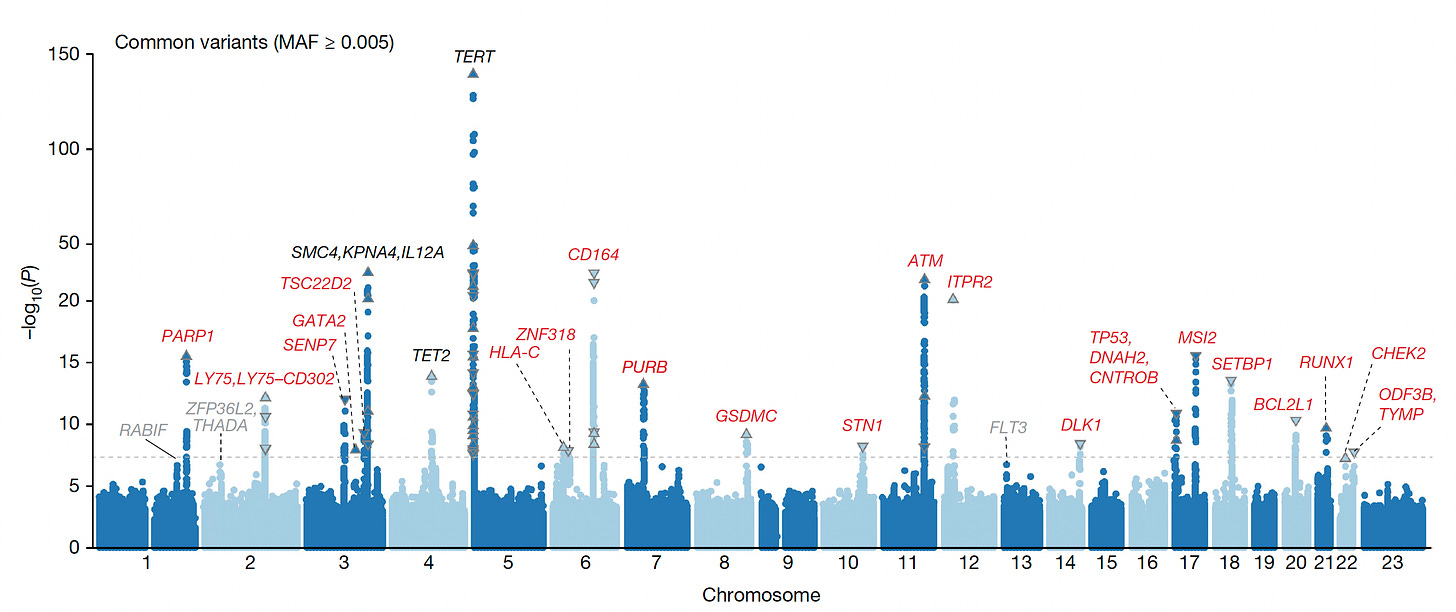
It is notable that CHIP is heritable from over 200,000 participants in the UK Biobank, and more recently from exome sequencing of over 620,000 people from Geisinger Health (with over 40,000 exhibiting CHIP), with the common genome loci associations in the Manhattan plot below, and a recent report of familial mutations in POT1 gene, which, interestingly, are part of a long telomere syndrome (in contrast to shortening of telomeres that are associated with aging), were linked with a variety of both benign and solid cancers. Key differences have been noted between the UK Biobank European and Japanese populations with respect to the implicated genome loci and mutations underpinning CHIP. The type of mutation—be it a single letter (SNP) or a structural variant—is also an important determinant of risk. Interestingly, last month, activation of a particular gene—TCL1A—was tied to developing CHIP—and a protective allele of this gene slowed HSC expansion, which could prove to be a tipoff for inhibiting CHIP in the future.
A Major Risk Factor for Heart Disease
As seen in the Table below, the independent hazard ratio (HR) risk for developing heart disease with CHIP is 1.8-fold, which is similar to type 2 diabetes (2.2- fold), worse than smoking, hypertension, and lipid abnormalities.
Multiple studies have reinforced the serious increased risk for cardiovascular outcomes, indicating the risk for early-onset heart attack is increased 4-fold compared with matched controls, and a two-fold increased risk of coronary heart disease. The associated risk of CHIP for developing heart failure, pulmonary embolism, deep vein thrombosis, atrial fibrillation, and stroke are all significantly increased. So is all-cause mortality risk. A simple schematic is shown below. Details of the multiple cardiovascular associations are provided in the excellent review by Tall and Fuster.
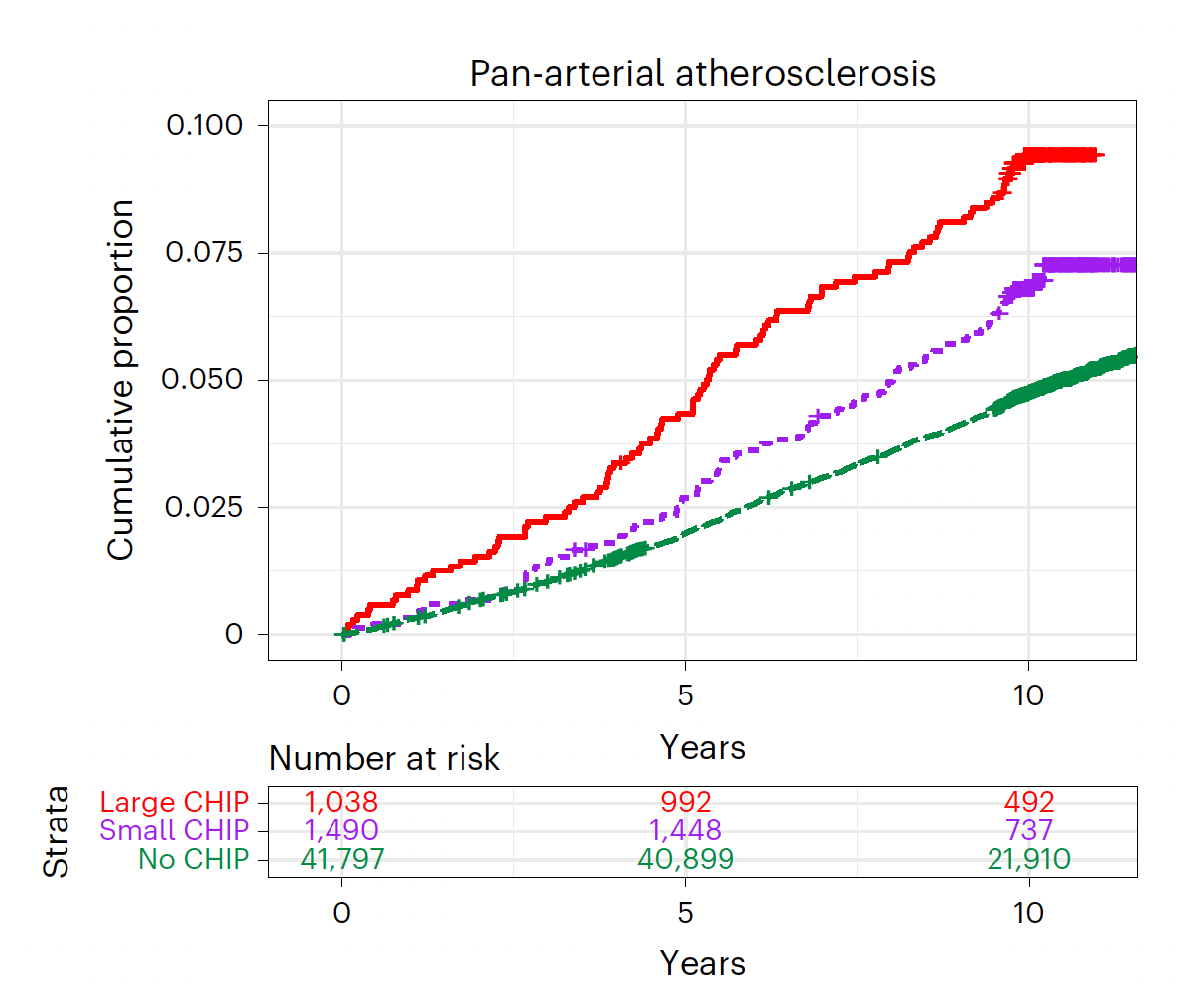
As seen below, the incidence of atherosclerosis across all arterial beds (coronary, cerebrovascular and peripheral) is increased proportionally with the extent of CHIPs
And, notably the relationship of type of CHIP mutations plotted for risk (hazard ratio) blood malignancy (AML=acute myelogenous leukemia) and for heart diseases (left panel below, CHD=coronary heart disease) or peripheral artery disease (right panel).
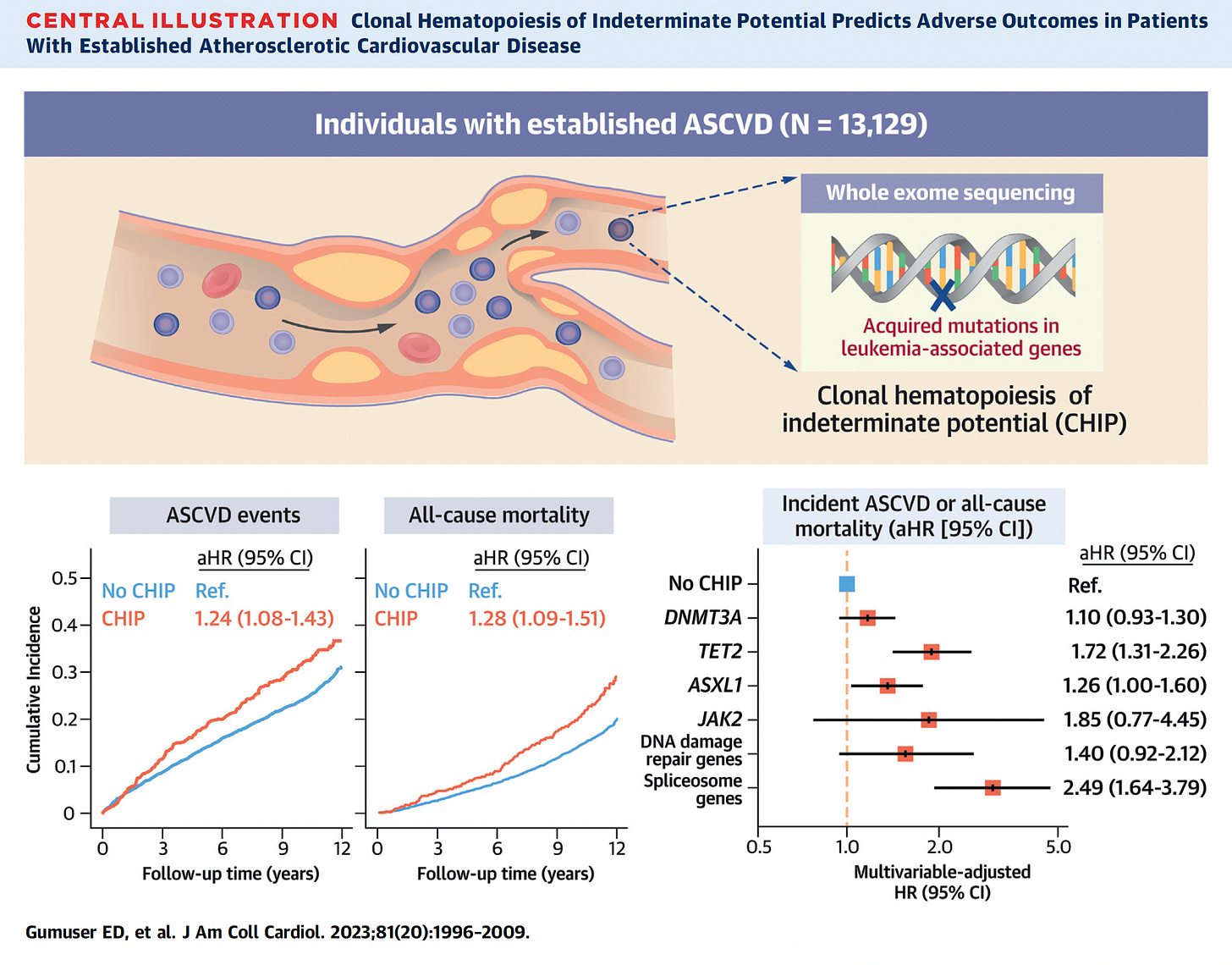
A publication from this week pulls together the risk of atherosclerotic events and all-cause mortality, along with the different CHIP mutation risks
There is a lingering question about CHIP and atherosclerosis like the chicken and the egg. It has been raised that atherosclerosis may be accelerating the risk of CHIP rather than a unidirectional cause and effect.
Chronic Liver Disease, Lung and Skin Cancer
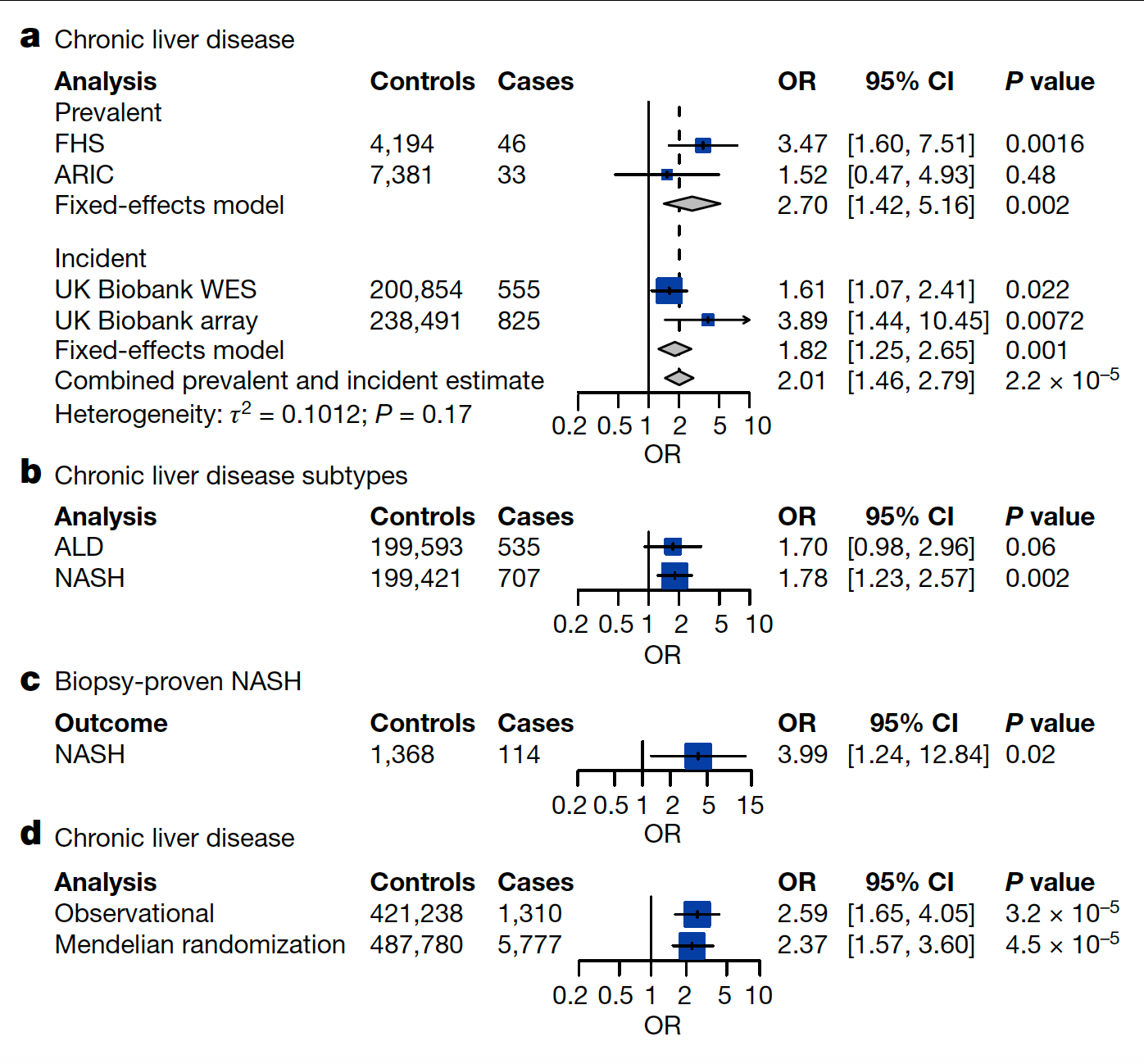
Well beyond blood malignancies, there is a heightened risk of various forms of chronic liver disease as seen in this graphic (OR-odds ratio).
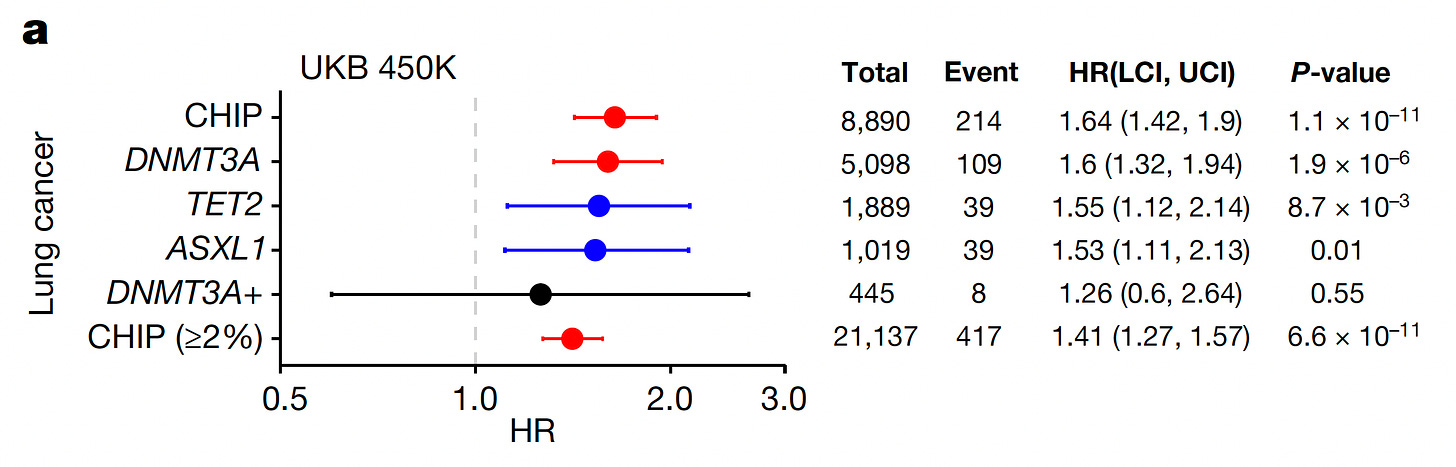
and lung cancer, as seen with the specific CHIP mutations below in the UK Biobank (UKB) and also for non-melanoma skin cancers (summarized here). Other studies have established a link to a variety of solid cancers and another form of clonal hematopoiesis called mosaic has been linked with sepsis, pneumonia and susceptibility to various types of infections.
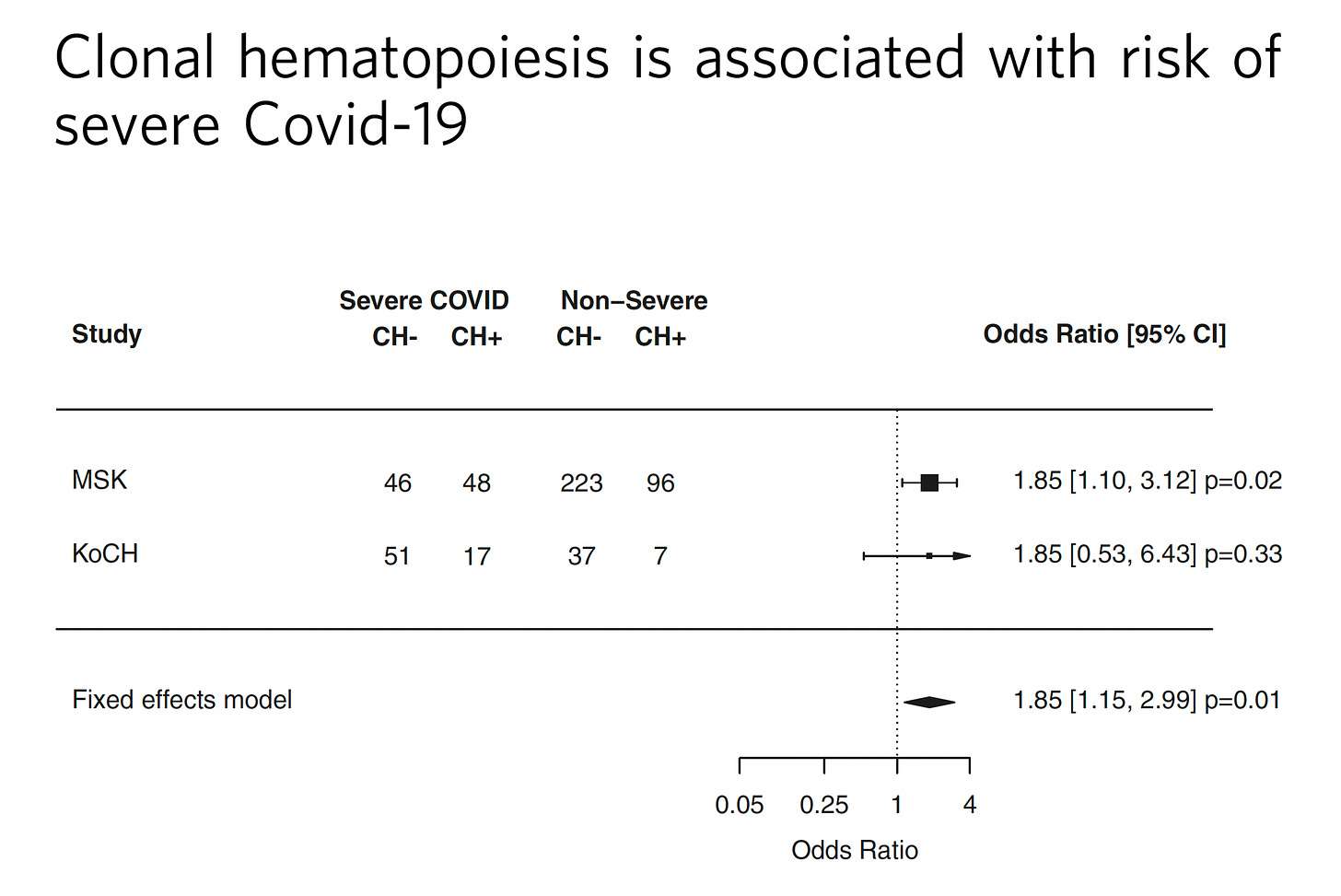
There’s even a link of CHIP to the risk of severe Covid, as was quickly pointed out from Leora Herrmann, a reader, after seeing this post.
Inflammaging and Treatment
The common thread of CHIP impact with aging, as verified in multiple experimental studies along with the body of clinical evidence I’ve summarized here, is heightened inflammation, what has been referred to as “inflammaging.” When the specific CHIP mutations are tested in animal models that promote inflammation and the process of atherosclerosis. The elevated pro-inflammatory cytokines (proteins secreted by cells) increase the likelihood of chronic diseases well beyond the well established blood malignancies linked to CHIP. The direct cause-and-effect of CHIP as pathogenic (disease-causing) is supported by these studies, but even if some of the effects are not directly attributable to CHIP, the utility of CHIP as an important biomarker has been firmly established.
With inflammation as the underpinning of CHIP’s trouble, why not test anti-inflammatory agents to determine whether we can suppress the adverse clinical outcomes? Very little work has been done to this end, with only one retrospective look at an interleukin monoclonal antibody (anti-IL-1β) called canakinumab in a subset of patients with CHIP TET2 variants (from a randomized trial) suggested potential for benefit, not seen in the subgroup of CHIP DNMT3A mutations, which at best can be considered quite preliminary. Added to this is the risk of this potent monoclonal antibody for susceptibility to infections. It is frankly surprising that much more work prospectively testing less potent anti-inflammatory agents has not been undertaken.
Without any validated treatments, many would conclude there is no reason to use this biomarker clinically. But that doesn’t compute.
A Call for CHIP Testing
As I started with this piece, the “IP” in CHIP stands for indeterminate potential. That’s wrong—it’s not “indeterminate.” It’s been determined that CHIP isn’t a good thing and carries risk, and that certain types of CHIP driver mutations are worse than others. And the more CHIP, the worse the outcomes. Yet clinically these hundreds of studies are all being ignored. No one gets a CHIP assessment as part of their medical workup. CHIP just sits in the medical research silo.
We clearly have a biomarker that is firmly validated as an important independent risk factor for significant cardiovascular outcomes, blood malignancies, no less many other major medical conditions. The main justification for its lack of clinical utility is that we don’t have a treatment. However, when we assess risk clinically we use factors that are not readily subject to change, like age, sex, diabetes, and hypertension. We’re now onto an era of multi-cancer early detection (MCED, reviewed previously) but we don’t factor in CHIP for who might benefit from these expensive screening tests. Or when we assess a person’s risk of heart or vascular disease. The time has come to ratchet up the testing of anti-inflammatory interventions to reduce the toll of CHIP, to determine ways that CHIP can be prevented or markedly delayed, and to consider ways to inexpensively assay the presence and type of CHIP in people of advanced age. At the very least, it will help us to determine which patients should have more close follow-up, without doing any additional testing that would promote incidentalomas and rabbit-hole workups. A chance to further work on known modifiable risk factors for a given individual with CHIP. With over a decade of solid work that has taught us about CHIP, it’s time to acknowledge there’s something there and get serious about it.
Thanks for reading and subscribing to Ground Truths. If you found this newsletter informative please share it.
Much gratitude to the paid subscribers who have helped fund many of our high school and college summer interns at Scripps Research.