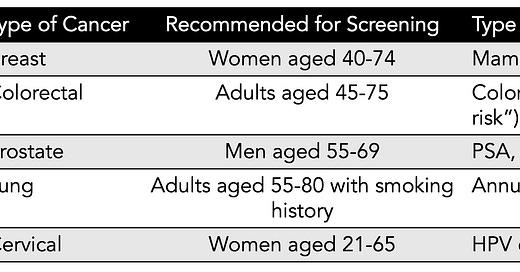
Age. It’s the one major criteria for cancer screening recommendations across all common cancers. The US Preventive Services Task Force (USPSTF) current recommendations are summarized below (simplified version, without grading of recommendation or details), with age as the basis for population-based screening. The cost of this mass, age-based screening per se, not including follow-up tests and procedures or over-diagnosis, is estimated to be between $40-80 billion per year. Yet, for example, 88% of women will never develop breast cancer in their lifetime, even though mammography is recommended every other year for women age 40 and older (as recently lowered from age 50 by USPSTF). Recently, these age cutoffs have been challenged with the emerging trend of cancer occurring in younger age groups. Adding to all of this is the approximate 10% false positive rate for many of the tests each time they are performed, such as mammography. For mammography, about half of women will have a false-positive test over the course of a decade (5 scans) of testing. Currently, in the United States, only 14% of diagnosed cancers are detected by screening with a recommended screening test.
Is this the best we can do?
We have a chance to radically improve the accuracy, efficiency, timeliness (early) of detection and cost-effectiveness for cancer screening by multiple means.
The “AI biopsy”
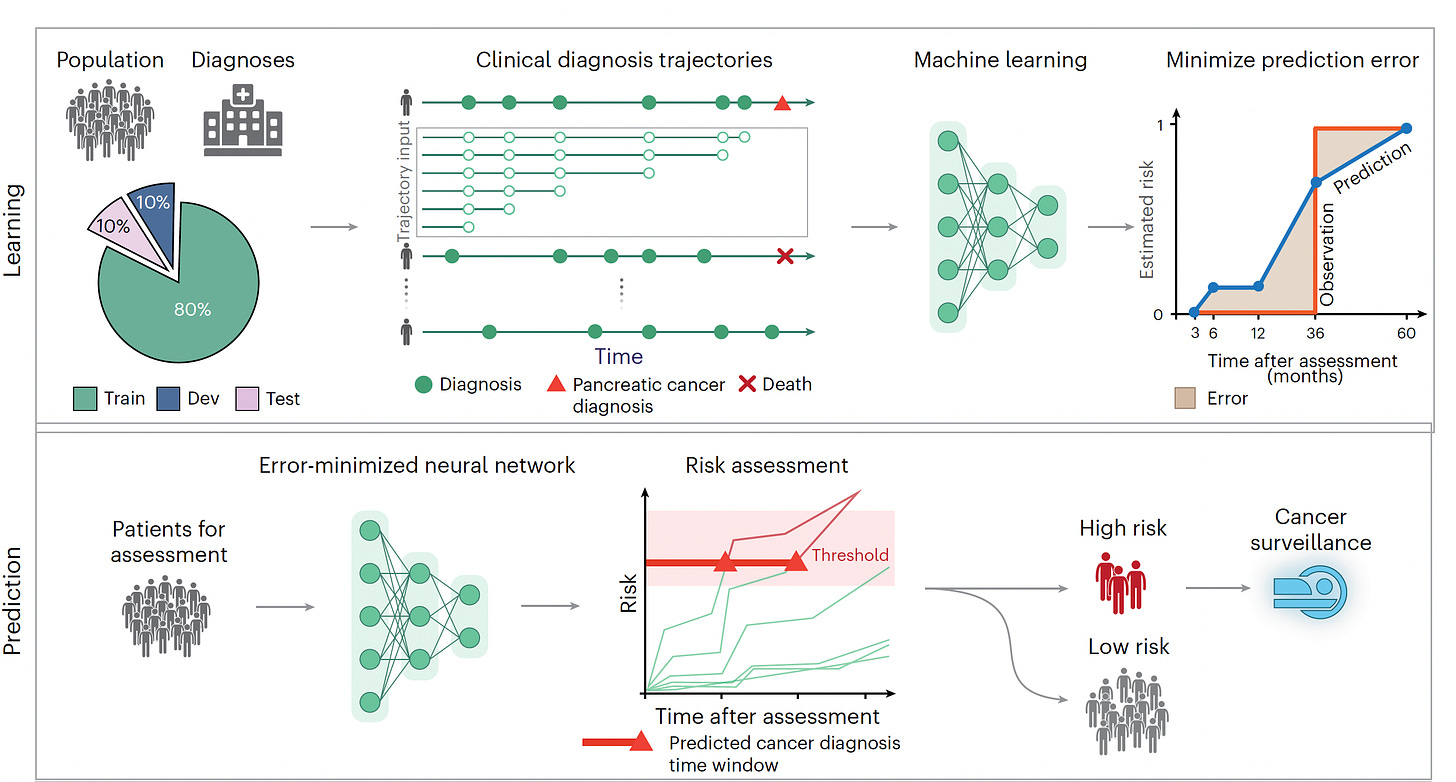
I recently reviewed the major study of AI for detecting pancreatic cancer using a transformer AI model in 2 large cohorts (Denmark and the US), in aggregate from more than 11 million people of whom 28,000 developed pancreatic cancer, one of the most deadly forms of cancer and exceedingly difficult to diagnose early. I won’t go through all the details again, but the A.I. performed well (AUC=0.88) that exceeded previous machine and deep learning models, and only incorporated minimal risk factor data derived from the history for each individual, with time-sequence data identified as particularly useful for aiding prediction. It did not include multiple other layers of data (“multimodal”), such as genomics, lab tests or scans.
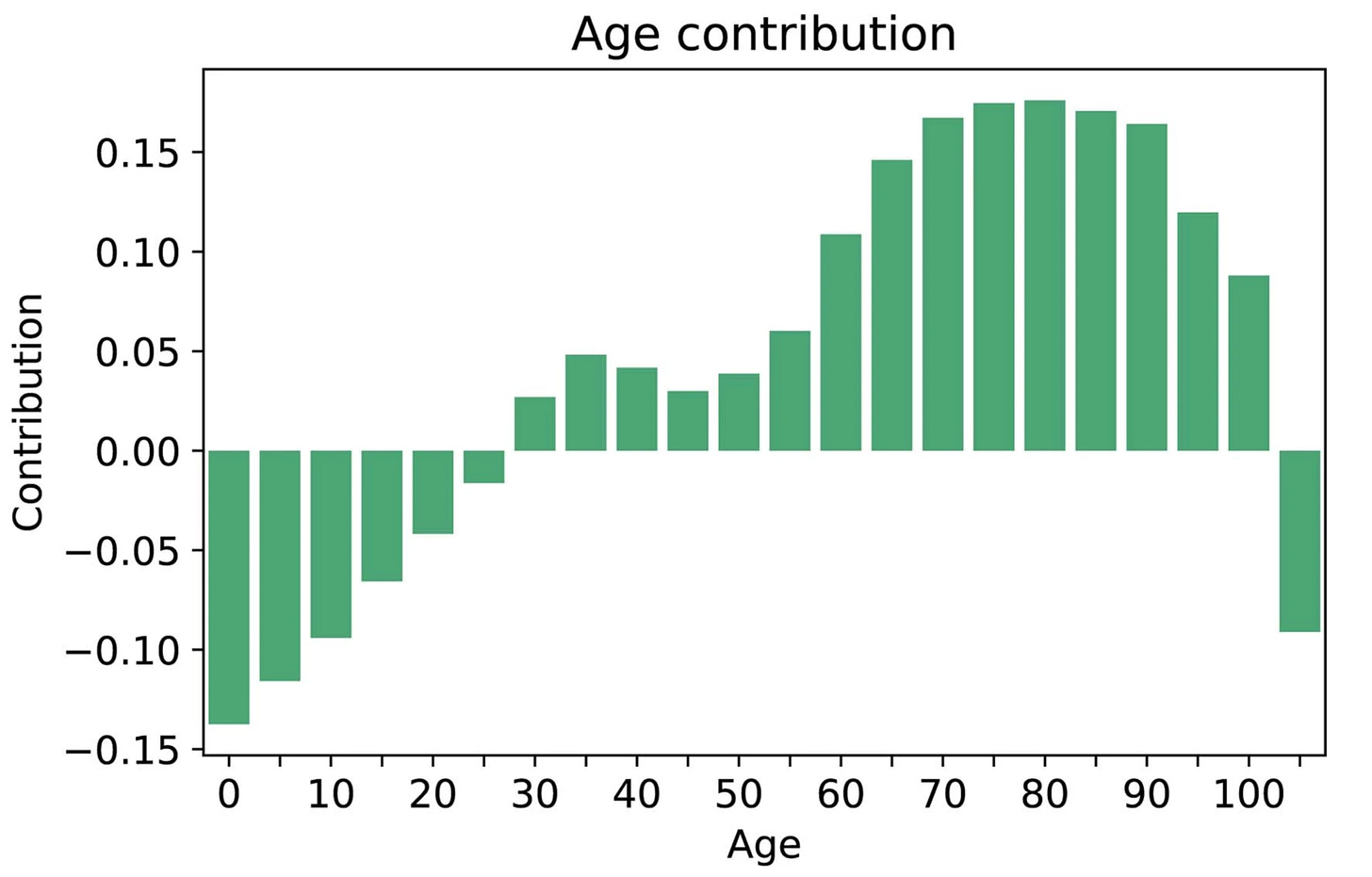
While age 50 plus was a key risk factor in the study, there was also a contribution of age for prediction in people ages 30 to 50 that would be missed using the strict age 50 cutoff.
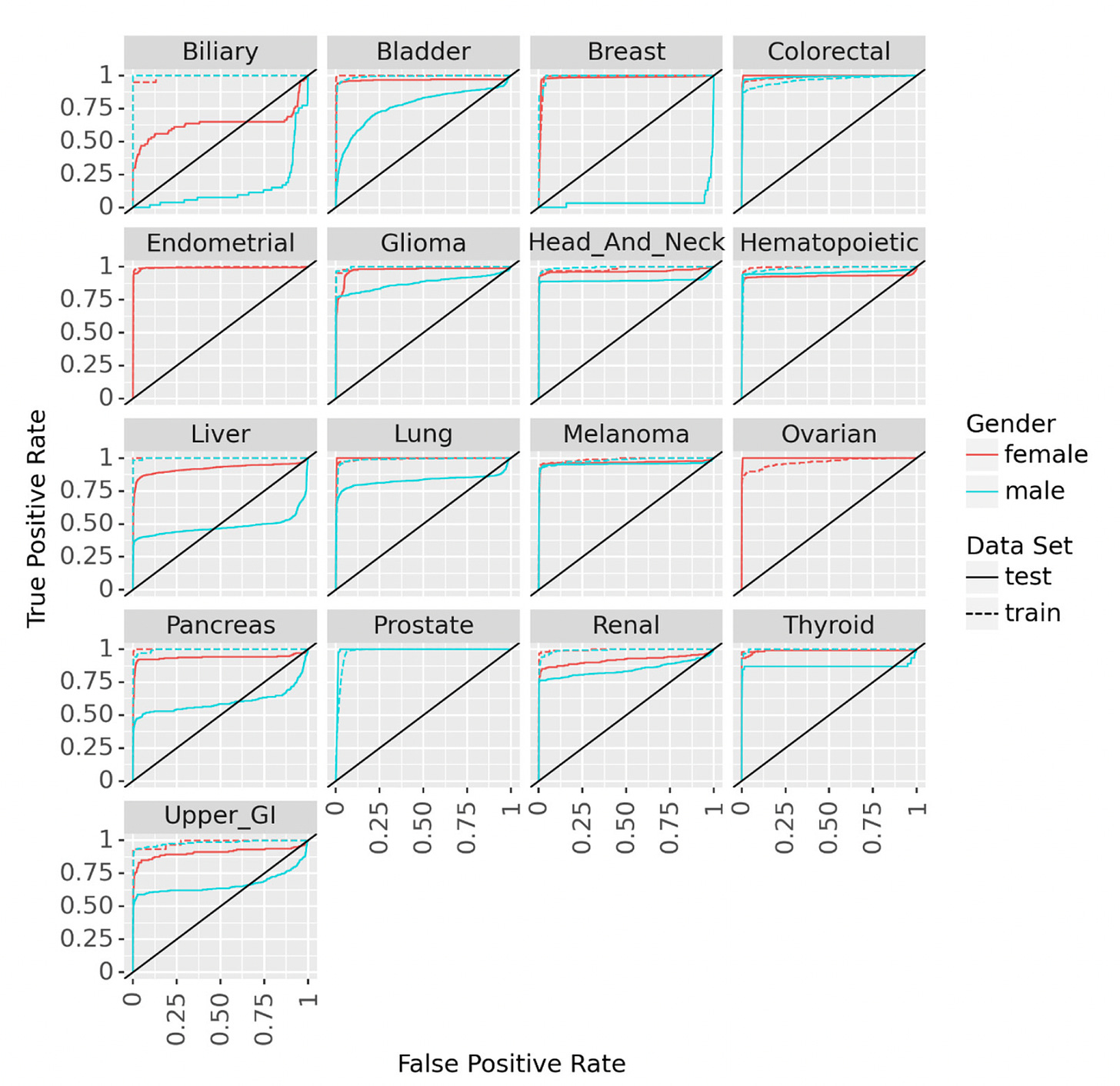
Hart and colleagues developed the term and wrote about a “statistical biopsy” using personal health data and two different deep neural networks for 17 types of cancer, assessed for the UK biobank cohort as seen below. Again, the data inputs were rather limited to demographics, medical, family and social history, without lab, scan, environmental or omic (such as genome sequence, gut microbiome, immune system profiling) data.
Individualized Genomic and Biologic Risk
Multiple tests that can indicate an individual’s heightened risk for developing cancer include polygenic risk scores, whole genome sequencing for mutations in cancer susceptibility genes, known pathogenic variants linked to cancers, gut microbiome profiles, clonal hematopoiesis of blood stem cells, and potentially immune system profiling. None of these are currently included to assess a person’s risk for cancer screening.
The least expensive of these tests are polygenic risk scores (PRS) which could be obtained from a gene microarray, such as used by 23andMe or AncestryDNA, which captures data for over 1 million common variants. Alternatively, low-pass whole genome sequencing can be used to derive polygenic risk scores. Both methods can be accomplished at very low cost, less than $50, and there are validated polygenic risk scores for most of the common cancers—prostate, breast, lung, colorectal and melanoma. There are recent data for how a PRS can be integrated with the prostate-specific antigen blood test to avoid 31% of negative prostate biopsies, and improve detection of aggressive cancer. A PRS for colorectal cancer was assessed in 400,000 Finnish individuals and found to identify people at high risk (top 1% score) who were younger than the population-based screening age (in Finland it is 60 years), concluding, at the European Society of Human Genetics in June 2023, “This indicates that a colorectal cancer-specific PRS would be able to define more appropriate ages to start screening for individuals based on their genetic risk.”
Whole genome sequencing, which can now be performed for about $200, provides data on the known pathogenic variants for familial cancers (such as BRCA and Lynch Syndrome) but also a detailed assessment for known cancer predisposition genes, that were established to exceed 100 a decade ago (Figure below). The good part about these types of genomic data is that they are a one-off, obtained one time to provide insight throughout a person’s life. Unfortunately, most of the validated datasets for genomic cancer risk are still for people of European ancestry.
There is also the biomarker of clonal hematopoiesis of indeterminate potential (CHIP) that I just reviewed, linked to heightened risk for a number of cancers including blood, lung and skin (it also predicts cardiovascular risk), that is not available commercially, but ideally should be. It can be a confounder for the MCED tests discussed below, but prospective assessment for partitioning risk should be feasible. There’s also the increased risk of cancer with specific gut microbiome profiles (Figure below) with decreasing diversity and reduced functional redundancy, along with a higher proportion of pathogenic microorganisms, and patterns specific to the different types of cancer, as seen below. This extends potentially to the oral microbiome as well. While the gut microbiome data can be inexpensively obtained, it isn’t yet being used for cancer risk assessment.
Like the microbiome, the individual’s immune system response, which is interdependent with the gut microbiome, and likely an important determinant for developing risk of cancer, but is not assessed. To be clear, I’m not advocating for the use of the gut or oral microbiome or immune profiling at this point, but they deserve attention as solid candidate ancillary tests to better define risk.
Multi-Cancer Early Detection (MCED) Blood Tests
In recent years, there has been intense interest for developing and validating a blood test (also known as “liquid biopsies”) to detect early cancer in healthy people, without any symptoms, to ultimately be incorporated in one’s annual checkup. The various tests assess patterns of methylation markers, DNA sequence, fragments of DNA, proteins, metabolites, exosomes, or RNA.
These tests are largely single-omic, that is they are each looking at one type of biologic marker. An exception is the one developed by Exact Sciences that includes some genomic and protein markers. It is quite likely that a multi-omic test, that included orthogonal (complementary) data across multiple types of biologic markers would be ideal, but that would bring the cost of the test to much higher than we have already seen, such as the Galleri test by Grail that costs $949 and that looks at the methylation pattern from the blood sample for over 50 types of cancer.
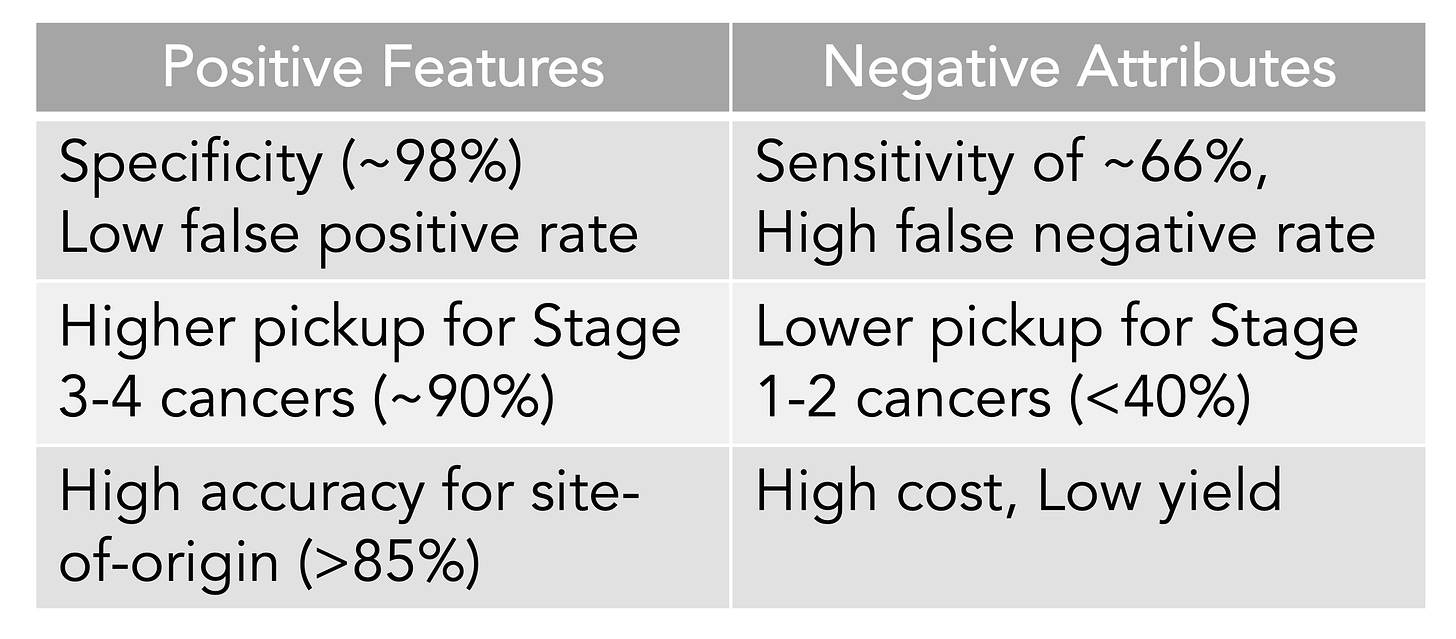
A meta-analysis of 10 case-control and 6 cohort studies for the different MCED tests came up with an overall sensitivity of 0.66 and a specificity of 0.98. That level of specificity— the true positive test—is the striking upside so far for the MCED tests, as shown below. The overall accuracy for predicting the cancer site of origin from the blood biomarker was 79%.
To date, the Galleri test has the most experience of the MCED tests with over 100,000 that have been done commercially and multiple reports, such as presented as the recent American Society of Clinical Oncology (ASCO) meeting or published. Simultaneously presented and published at ASCO was the SYMPLIFY study performed by the National Health Service in the UK of over 6,000 enrolled participants for whom 6.7% had cancer diagnosed (5,461 had the Grail test, of whom 368 had a cancer diagnosis). Importantly, this first large-scale prospective study included people with symptoms, but the sensitivity and specificity were very closely aligned with the meta-analysis of asymptomatic people, 66% and 98%, respectively. An impressive result for this study was the prediction of the cancer site-of-origin which uses machine learning from the methylation pattern and was accurate in 85%.
That accuracy for determination of site of origin was replicated in another report for the Galleri test at ASCO from the PATHFINDER trial which also assessed over 6,600 participants age 50 plus, but without symptoms. A cancer signal was found in 92 participants (1.4%) of whom 35 (0.5%) were true positives, for a specificity of 99% (35 of 36 found to have cancer) but a positive predictive value of only 38%. The cancer site of origin prediction accuracy was 97%. Given the diverse sites of origin, including bone, head and neck, ovary, liver, uterus, blood, breast, lung, and various GI tract, that’s quite good.
Of note, there were 57 false positives, participants that had no findings of cancer after a full workup. What remains to be seen is whether the signal of abnormal cell-free DNA methylation is actually a “true” positive in some individuals, such that the person’s immune system kicks in and, on repeat assessment months later, the test comes back negative. That some individuals have microscopic cancer and then their immune system kicks in to quash it. There are anecdotal reports of this occurrence which will require further scrutiny.
The Table below is my summary of what we know about MCED tests from the meta-analysis and the new Galleri data from the ASCO meeting and publications.
The detection rate for early cancer (stage 1 and 2) is clearly suboptimal compared with later stages, which is a premise for MCED—that picking up cancer at the microscopic stage, before it can be seen on a scan or induce symptoms, or potentiate spread, is the key to improving outcomes, using interventions to change the natural history of cancer progression. Simply put, detection is not equivalent to favorably altering outcomes. That is why randomized trials are necessary and ongoing for MCED vs no MCED for clinical outcomes, such as one by the National Health Service in the UK of 140,000 participants (now fully enrolled in just 10 months) and the National Cancer Institute Vanguard study plan in preparation for a potential 225,000 person trial using a MCED derived on their own. These very large trials of healthy, asymptomatic people are again using age as the main inclusion criteria. It will be years before such trials will be complete for clinical outcomes, and there will be questions about the diversity of the participants, since we know the occurrence and behavior of many cancers are shaped by ancestry background. But the problem of lacking compelling evidence for MCED benefit from randomized trials is only one of the critical issues. None of the tests are FDA approved or reimbursed by insurance carriers. Let’s now get into the bigger problem.
A Detection Yield of 5 per 1000 People Tested Won’t Cut It
The basic problem with MCEDs is that they are primarily being assessed by the same dumbed-down criteria of current mass screening—age 50 plus. This ignores the Bayesian principle of priors—that using a test for low-risk individuals will lead to false positives and low predictive value. Indeed, in PATHFINDER there were more false positives than true positives (57 vs 35) and the signal rate in 92 people of over 6,600 assessed is dreadfully low. While remarkably specific, the low sensitivity leaves dangling the number of people who may have microscopic cancer but are not being detected. Any medical test will only be as good for accuracy as the population it is tested in. There is a dire need to improve the signal and decrease the noise.
There are 2 major ways to improve the accuracy. One would be to enrich the population to be tested, using the “AI biopsy” described above with all of the longitudinal clinical information, beyond that which was used for the pancreatic cancer detections study. That would be comprehensive: inclusive of analyzing trends in lab data even within the normal range, all scans, nutrition, socioeconomic features, and environmental exposures such as air pollution. Added to this would be individualized genomic/biologic risk determination. Now that we have large language models with multimodal input capabilities, the analytic challenge is clearly surmountable. Then we have defined a high-risk population that may derive marked benefit from an MCED test.
To get at early detection at the Stage 1 level, it may be important to have multi-omic MCEDs, not just one focused on methylation or DNA sequence. But, as discussed above, this may make the cost prohibitive. It may well be that enriching the population tested is the driver towards higher sensitivity, without the need for multi-omic assessment, the trade-offs of the individual’s genomic assessment vs more omics of the blood sample deserves consideration.
For now, MCEDs are a test for the curious affluent that only carry potential value if positive, and only about half of those will be indicative of an actual cancer diagnosis that was not otherwise apparent. It will be primarily used by the same people who get executive health physicals with all sorts of tests that have not been validated in healthy people, at exorbitant costs.
If, however, we adopt a smarter path for assessing MCEDs ,they may indeed fulfill their transformative potential of being part of an annual physical—perhaps even eliminating the mass screening procedural tests as we know them today, such as mammography and colonoscopy.
In 2023, more than 600,000 Americans will die of cancer and about 2 million will have a new diagnosis of cancer, many at late stages. We have the desperate need to improve upon these statistics, and, through generative A.I. and biomedical innovation, we now have the tools to achieve this big unmet goal. We have come a long way for understanding a healthy individual’s risk of cancer and interpreting the molecular biologic content of a blood sample. If only we can get smart about using all this information.
Thanks for reading Ground Truths!
Please share this piece if you found it helpful.
I’m appreciative of your free subscription. If you decide to become a paid subscriber, know that all proceeds go to Scripps Research and have already been used to support many of our high school and college summer interns.