Full videos of all Ground Truths podcasts can be seen on YouTube here. The audios are also available on Apple and Spotify.
Transcript with audio and external links
Eric Topol (00:05):
Hello, it's Eric Topol with Ground Truths, and I am really thrilled to have with me Professor Faisal Mahmood, who is lighting it up in the field of pathology with AI. He is on the faculty at Harvard Medical School, also a pathologist at Mass General Brigham and with the Broad Institute, and he has been publishing at a pace that I just can't believe we're going to review that in chronological order. So welcome, Faisal.
Faisal Mahmood (00:37):
Thanks so much for having me, Eric. I do want to mention I'm not a pathologist. My background is in biomedical imaging and computer science. But yeah, I work very closely with pathologists, both at Mass General and at the Brigham.
Eric Topol (00:51):
Okay. Well, you know so much about pathology. I just assume that you were actually, but you are taking computational biology to new levels and you're in the pathology department at Harvard, I take it, right?
Faisal Mahmood (01:08):
Yeah, I'm at the pathology department at Mass General Brigham. So the two hospitals are now integrated, so I'm at the joint department.
Eric Topol (01:19):
Good. Okay. Well, I'm glad to clarify that because as far as I knew you were hardcore pathologist, so you're changing the field in a way that is quite unique, I should say, because a number of years ago, deep learning was starting to get applied to pathology just like it was and radiology and ophthalmology. And we saw some early studies with deep learning whereby you could find so much more on a slide that otherwise would be not even looked at or considered or even that humans wouldn't be able to see. So maybe you could just take us back first to the deep learning phase before these foundation models that you've been building, just to give us a flavor for what was the warmup in this field?
Faisal Mahmood (02:13):
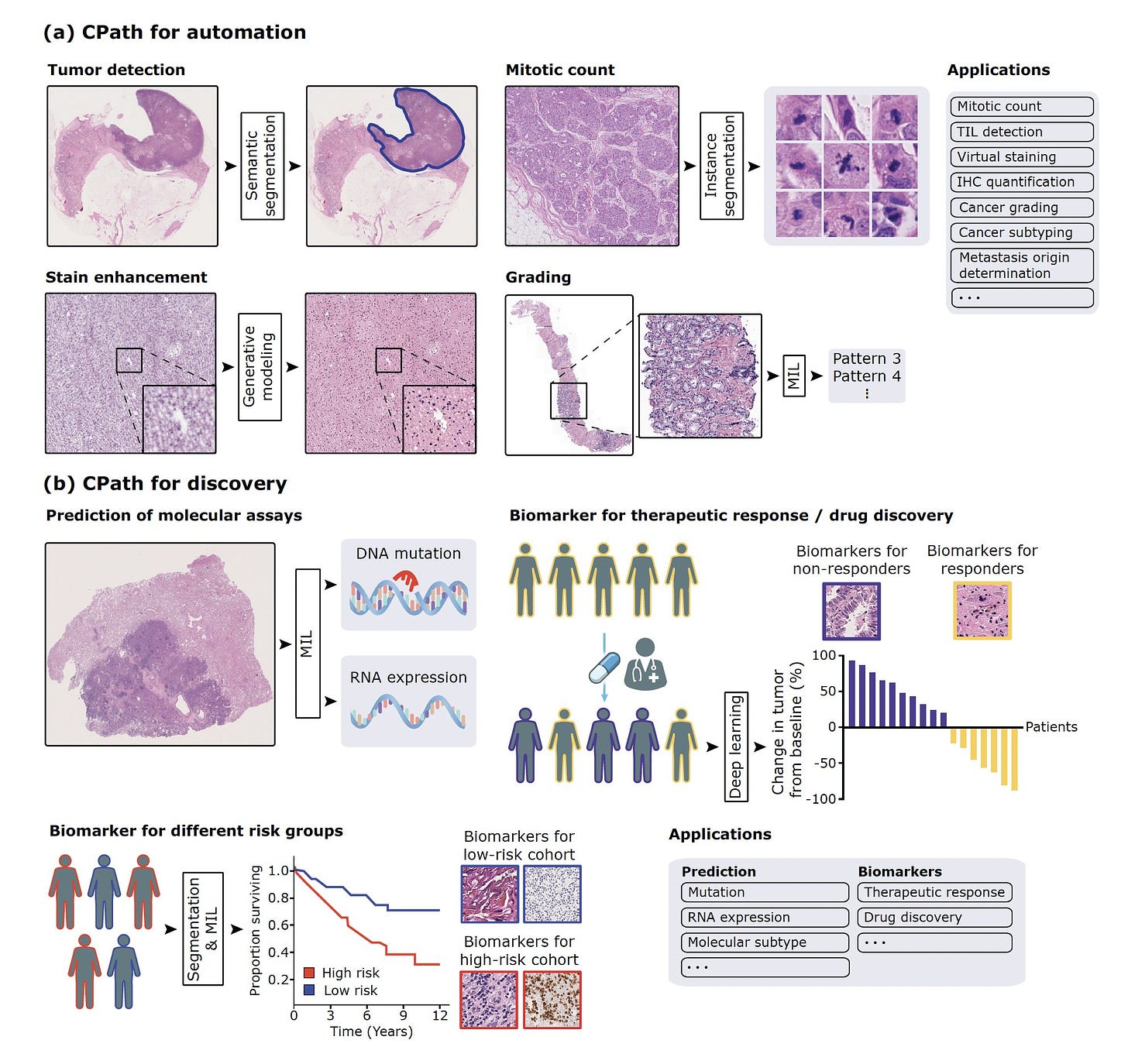
Yeah, so I think around 2016 and 2017, it was very clear to the computer vision community that deep learning was really the state of the art where you could have abstract feature representations that were rich enough to solve some of these fundamental classification problems in conventional vision. And that's around the time when deep learning started to be applied to everything in medicine, including pathology. So we saw some earlier cities in 2016 and 2017, mostly in machine learning conferences, applying this to very basic patch level pathology dataset. So then in 2018 and 2019, there were some studies in major journals including in Nature Medicine, showing that you could take large amounts of pathology data and classify what's known to us and including predicting what's now commonly referred to as non-human identifiable features where you could take a label and this could come from molecular data, other kinds of data like treatment response and so forth, and use that label to classify these images as responders versus non-responders or having a certain kind of mutation or not.
(03:34):
And what that does is that if there is a morphologic signal within the image, it would pick up on that morphologic signal even though humans may not have picked up on it. So it was a very exciting time of developing all of these supervised, supervised foundation models. And then I started working in this area around 2019, and one of the first studies we did was to try to see if we can make this a little bit more data efficient. And that's the CLAM method that we published in 2021. And then we took that method and applied it to the problem of cancers of unknown primary, that was also in 2021.
Eric Topol (04:17):
So just to review, in the phase of deep learning, which was largely we're talking about supervised with ground truth images, there already was a sign that you could pick up things like the driver mutation, the prognosis of the patient from the slide, you could structural variations, the origin of the tumor, things that would never have been conceived as a pathologist. Now with that, I guess the question is, was all this confined to whole slide imaging or could you somehow take an H&E slide conventional slide and be able to do these things without having to have a whole slide image?
Faisal Mahmood (05:05):
So at the time, most of the work was done on slides that were fully digital. So taking a slide and then digitizing the image and creating a whole slide image. But we did show in 2021 that you could put the slide under a microscope and then just capture it with a camera or just with a cell phone coupled to a camera, and then still make those predictions. So these models were quite robust to that kind of domain adaptation. And still I think that even today the slide digitization rate in the US remains at around 4%, and the standard of care is just looking at a glass light under a microscope. So it's very important to see how we can further democratize these models by just using the microscope, because most microscopes that pathologists use do have a camera attached to them. So can we somehow leverage that camera to just use a model that might be trained on a whole slide image, still work with the slide under a microscope?
Eric Topol (06:12):
Well, what you just said is actually a profound point that is only 4% of the slides are being reviewed digitally, and that means that we're still an old pathology era without the enlightenment of machine eyes. I mean these digital eyes that can be trained even without supervised learning as we'll get to see things that we'll never see. And to make, and I know we'll be recalling back in 2022, you and I wrote a Lancet piece about the work that you had done, which is very exciting with cardiac biopsies to detect whether a heart transplant was a rejection. This is a matter of life or death because you have to give more immunosuppression drugs if it's a rejection. But if you do that and it's not a rejection or you miss it, and there's lots of disagreement among pathologists, cardiac pathologists, regarding whether there's a transplant. So you had done some early work back then, and because much of what we're going to talk about, I think relates more to cancer, but it's across the board in pathology. Can you talk about the inner observer variability of pathologists when they look at regular slides?
Faisal Mahmood (07:36):
Yeah. So when I first started working in this field, my kind of thinking was that the slide digitization rate is very low. So how do we get people to embrace and adapt digital pathology and machine learning models that are trained on digital data if the data is not routinely digitized? So one of my kind of line of thinking was that if we focus on problems that are inherently so difficult that there isn't a good solution for them currently, and machine learning provides, or deep learning provides a tangible solution, people will be kind of forced to use these models. So along those lines, we started focusing on the cancers of unknown primary problem and the myocardial biopsy problem. So we know that the Cohen’s kappa or the intra-observer variability that also takes into account agreement by chance is around 0.22. So it's very, very low for endomyocardial biopsies. So that just means that there are a large number of patients who have a diagnosis that other pathologists might not agree with, and the downstream treatment regimen that's given is entirely based on that diagnosis. The same patient being diagnosed by a different cardiac pathologist could be receiving a very different regimen and could have a very, very different outcome.
(09:14):
So the goal for that study is published in Nature of Medicine in 2022, was to see if we could use deep learning to standardize that and have it act as an assistive tool for cardiac pathologists and whether they give more standardized responses when they're given a machine learning based response. So that's what we showed, and it was a pleasure to write that corresponding piece with you in the Lancet.
Eric Topol (09:43):
Yeah, no, I mean I think that was two years ago and so much has happened since then. So now I want to get into this. You've been on a tear every month publishing major papers and leading journals, and I want to just go back to March and we'll talk about April, May, and June. So back in March, you published two foundation models, UNI and CONCH, I believe, both of these and back-to-back papers in Nature Medicine. And so, maybe first if you could explain the foundation model, the principle, how that's different than the deep learning network in terms of transformers and also what these two different, these were mega models that you built, how they contributed to help advance the field.
Faisal Mahmood (10:37):
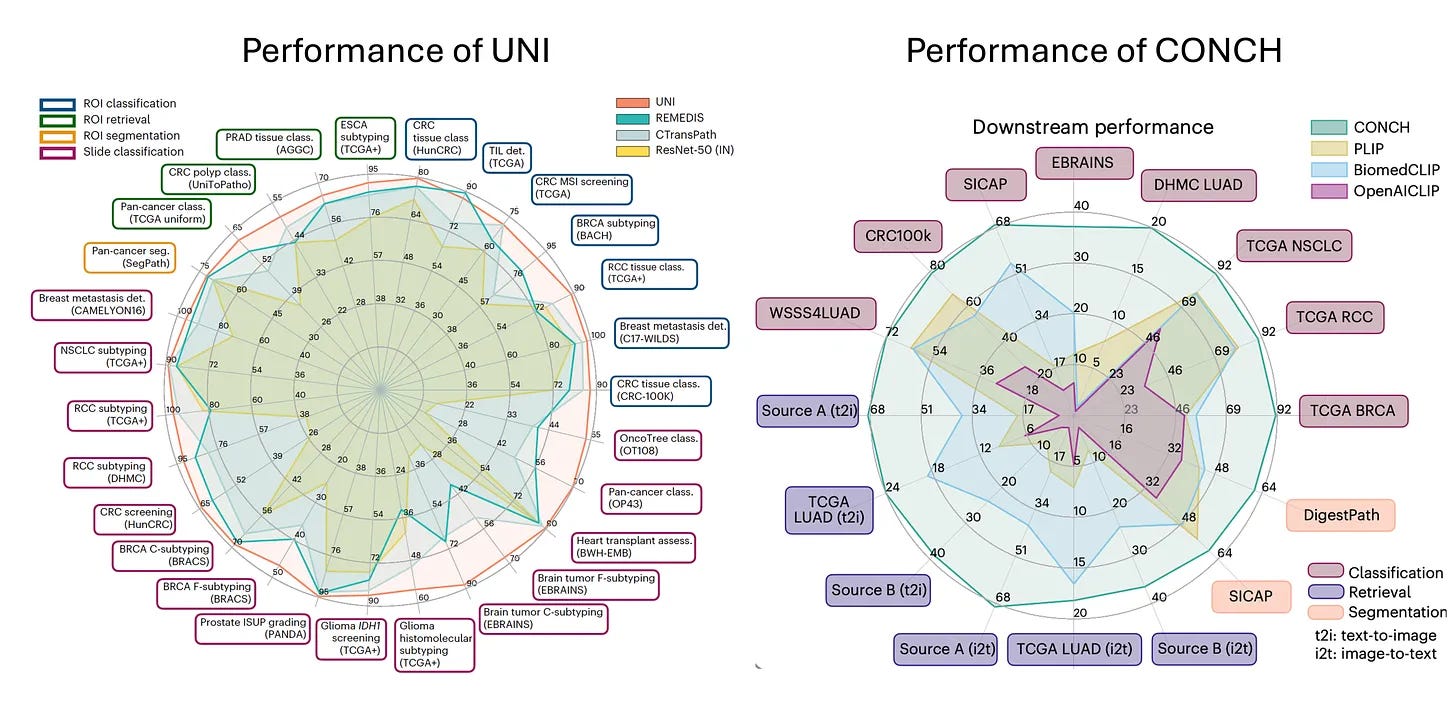
So a lot of the early work that we did relied on extracting features from a resonant trained on real world images. So by having these features extracted, we didn't need to train these models end to end and allowed us to train a lot of models and investigate a lot of different aspects. But those features that we used were still based on real world images. What foundation models led us do is they leveraged self supervised learning and large amounts of data that would be essentially unlabeled to extract rich feature representations from pathology images that can then be used for a variety of different downstream tasks. So we basically collected as much data as we could from the Brigham and MGH and some public sources while trying to keep it as diverse as possible. So the goal was to include infectious, inflammatory, neoplastic all everything across the pathology department while still being as diverse as possible, including normal tissue, everything.
(11:52):
And the hypothesis there, and that's been just recently confirmed that the hypothesis was that diversity would matter much more than the quantity of data. So if you have lots and lots of screening biopsies and you use all of them to train the foundation model, there isn't enough diversity there that it would begin to learn those fundamental feature representations that you would want it to learn. So we used all of this data and then trained the UNI model and then together with it was an image text model where it starts with UNI and then reinforces the feature representations using images and texts. And that sort of mimics how humans learn about pathology. So a new resident, new trainee learning pathology has a lot of knowledge of the world, but it's perhaps looking at a pathology image for the first time. But besides looking at the image, they're also being reinforced by all these language cues from, whether it's from text or from audio signals. So the hope there was that text would kind of reinforce that and generate better feature representation. So the two studies were made available together. They were published in Nature Medicine back in March, and with that we made both those models public. So at the time we obviously had no idea that they would generate so much interest in this field, downloaded 350,000 times on Hugging Face and used for all kinds of different applications that I would've never thought of. So that's been very exciting to see.
Eric Topol (13:29):
Can you give some examples of some of the things you wouldn't have thought of? Because it seems like you think of everything.
Faisal Mahmood (13:35):
Yeah, people have used it to when there was a challenge for detecting tuberculosis, I think in a very, very different kind of a dataset. It was from the Nightingale Foundation and they have large data sets. So that was very interesting to see. People have used it to create newer data sets that can then be used for training additional foundation models. It's being used to extract rich feature representations from pathology images, corresponding spatial transcriptomic data, trying to predict spatial transcriptomics directly from histology. And there's a number of other options.
Eric Topol (14:27):
Well, yeah, that was March. Before we get to April, you slipped in the spatial omics thing, which is a big deal that is ability to look at tissue, human tissue over time and space. I mean the spatial temporal, it will tell us so much whether an evolution of a cancer process or so many things. Can you just comment because this is one of the major parts of this new era of applying AI to biology?
Faisal Mahmood (15:05):
So I think there are a number of things we can do if we have spatial data spatially resolved omic data with histology images. So the first thing that comes to my mind as a computer scientist would be that can we train a joint foundation model where we would use the spatially resolved transcriptomics to further enforce the pathology signal as a ground truth in a contrastive manner, similar to what we do with text, and can we use that to extract even richer feature representation? So we're doing that. In fact, we made a data set of about a thousand pathology images with corresponding spatial transcriptomic information, both curated from public resources as well as some internal data publicly available so people could investigate that question further. We're entrusted in other aspects of this because there is some indication including a study from James Zou’s group at Stanford showing that we can predict histology, predict the spatial transcriptomic signal directly from histology. So there's early indications that we might also be able to do that in three dimensions. So yeah, it's definitely very interesting. More and more of that data is becoming available and how machine learning can sort of augment that is very exciting.
Eric Topol (16:37):
Yeah, I mean, most of the spatial omics has been a product of single cell sequencing, whether it's single nuclei and different omics, not just DNA, of course, RNA and even methylation, whatnot. So the fact that you could try to impute that from the histologies is pretty striking. Now, that was March and then in April you published to me an extraordinary paper about demographic bias and how generative AI, we're in the generative AI year now since as we discussed with foundation models, here again that gen AI could actually reduce biases and enhance fairness, which of course is so counterintuitive to everything that's been written to date. So maybe you can take us through how we can get a reduction in bias in pathology.
Faisal Mahmood (17:34):
Yeah, so in the study, the study was about, this had been investigated in other fields, but what we try to show is that a model trained on large, diverse, publicly available data. When that's applied internally and we stratify it based on demographic differences, race and so forth, we see these very clear disparities and biases. And we investigated a lot of different solutions that were out there to equalize the distribution of the data to balance the distribution using or sampling and some of these simple techniques. And none of them worked quite well. And then we observed that using foundation models or just having richer feature representations eliminates some of those biases. In parallel, there was another study from Google where they use generative AI to synthesize additional images from those underrepresented groups and then use those images to enhance the training signal. And then they also showed that you could reduce those biases.
(18:49):
So I think the common denominator there is that richer feature representations contribute to reduced biases. So the biases not because there is some inherent signal tied to these subgroups, but the bias is essentially there because the feature representations are not strong enough. Another general observation is that there's some kind of a confounder often there that leads to the bias. And one example would be that patients with socioeconomic disparities might just be diagnosed late and there might not be enough advanced cases in the training dataset. So quite often when you go in and look at what your training distribution looks like and how it varies from your test distribution and what that dataset shift is, you're able to figure out where the bias inherently comes from. But as a general principle, if you use the richest possible feature representation or focus on making your feature representations richer by using better foundation models and so forth, you are able to reduce a lot of the bias.
Eric Topol (19:58):
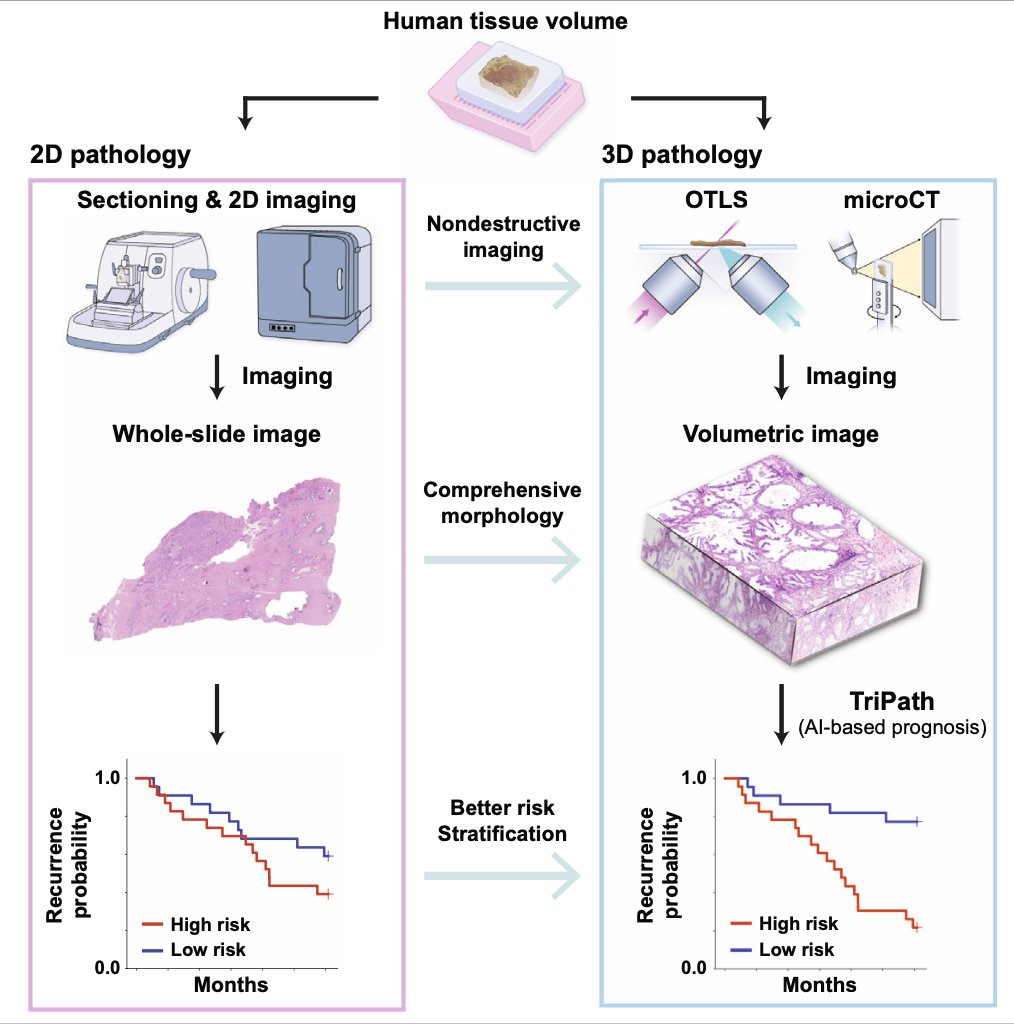
Yeah, that's really another key point here is about the richer features and the ability counterintuitively to actually reduce bias. And what is important in interrogating data inputs, as you said before, you wind up with a problem with bias. Now, then it comes May since we're just March and April, in May you published TriPath, which is now bringing in the 3D world of pathology. So maybe you can give us a little skinny on that one.
Faisal Mahmood (20:36):
Yeah. So just looking at the spectrum of where pathology is today, I think that we all agree in the community that pathologists often look at extremely sampled tissue. So human tissue is inherently three-dimensional, and by the time it gets to a pathologist, it's been sampled and cut so many times that it often would lack that signal. And there are a number of studies that have shown that if you subsequently cut sections, you get to a different outcome. If you look at multiple slides for a prostate biopsy, you get to a different Gleason score. There are all of these studies that have shown that 3D pathology is important. And with that, there's been a growing effort to build tools, microscopes, imaging tools that can image tissue in 3D. And there are about 10 startups who've built all these different technologies, open-top light-sheet microscopy, microCT and so forth that can image tissue really well in three dimensions, but none of them have had clinical adoption.
(21:39):
And we think that a key reason is that there isn't a good way for a pathologist to examine such a large volume of tissue. If they spend so much time examining this large volume of tissue, they would never be able to get through all the, so the goal here really was to develop a computational tool that would look through the large volume and highlight key regions that a pathologist can then examine. And the secondary goal was that does using three dimensional tissue actually improve patient stratification and does using, essentially using three 3D deep learning, having 3D convolutions extract richer features from the three dimensions that can then be used to separate patients into distinct risk groups. So that's what we did in this particular case. The study relied on a lot of data from Jonathan Liu's group at University of Washington, and also data that we collected at Harvard from tissue that came from the Brigham and Women's Hospital. So it was very exciting to show that what the value of 3D pathology can be and how it can actually translate into the clinic using some of these computational tools.
Eric Topol (22:58):
Do you think ultimately someday that will be the standard that you'll have a 3D assessment of a biopsy sample?
Faisal Mahmood (23:06):
Yeah, I'm really convinced that ultimately 3D would become the standard because the technology to image these tissue is becoming better and better every year, and it's getting closer to a point where the imaging can be fast enough to get to clinical deployment. And then on the computational end, we're increasingly making a lot of progress.
Eric Topol (23:32):
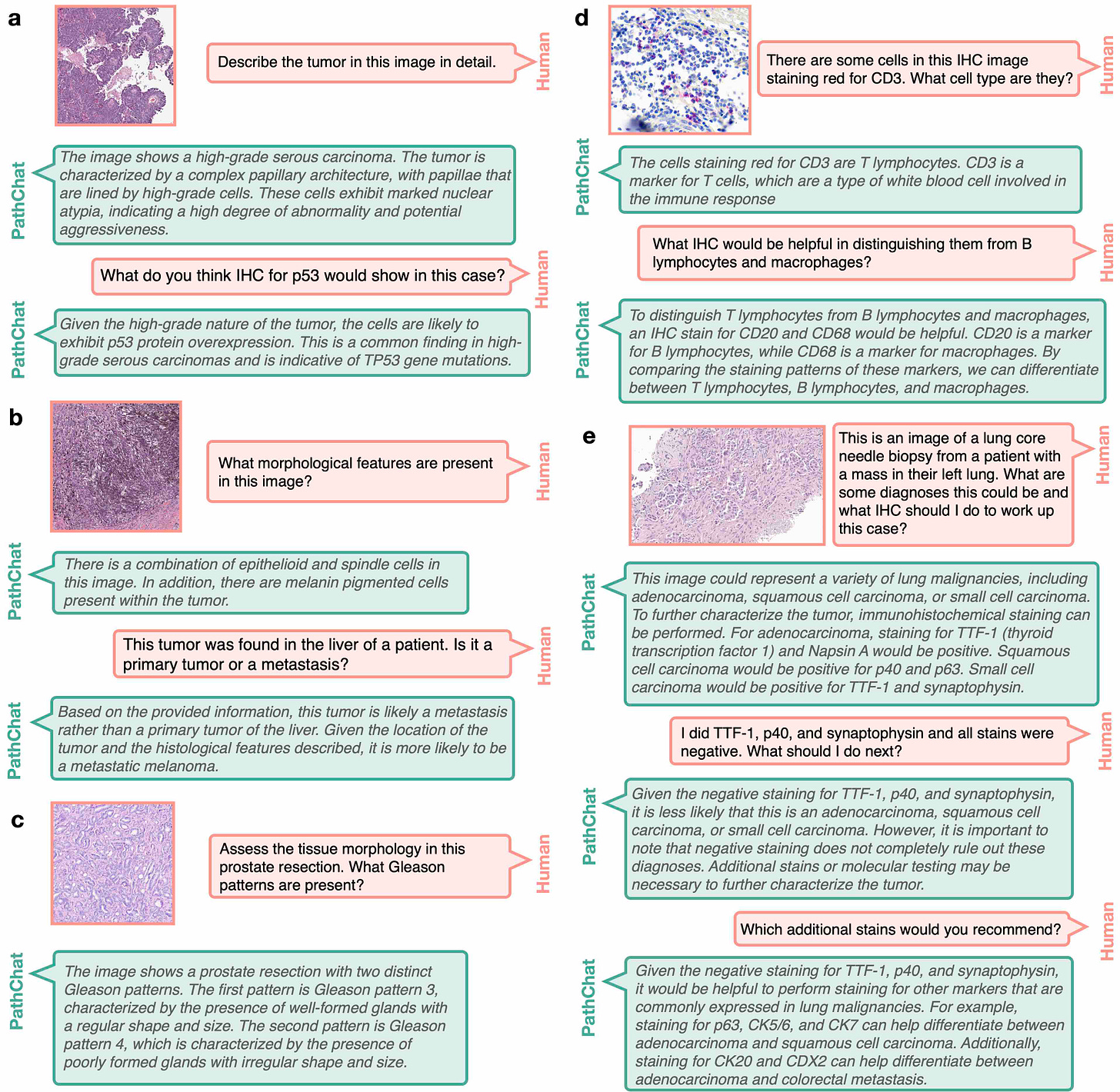
And it seems, again, it's something that human eyes couldn't do because you'd have to look at hundreds of slides to try to get some loose sense of what's going on in a 3D piece of tissue. Whereas here you're again taking advantage, exploiting the digital eyes. Now this culminates to your June big paper PathChat in Nature, and this was a culmination of a lot of work you've been doing. I don't know if you do any sleep or your team, but then you published a really landmark paper. Can you take us through that?
Faisal Mahmood (24:12):
Yeah, so I think that with the foundation models, we could extract very rich feature representation. So to us, the obvious next step was to take those feature representations and link them with language. So a human would start to communicate with a generative AI model where we could ask questions about what's going on in a pathology image, it would be capable of making a diagnosis, it would be capable of writing a report, all of those things. And the reason we thought that this was really possible is because pathology knowledge is a subset of the world's knowledge. And companies like OpenAI are trying to build singular, multimodal, large language models that would harbor the world's information, the world knowledge and pathology is much, much more finite. And if we have the right kind of training data, we should be able to build a multimodal large language model that given any pathology image, it can interpret what's going on in the image, it can make a diagnosis, it can run through grading, prognosis, everything that's currently done, but also be an assistant for research, analyzing lots of images to see if there's anything common across them, cohorts of responders versus non-responders and so forth.
(25:35):
So we started by collecting a lot of instruction data. So we started with the foundation models. We had strong pathology image foundation models, and then we collected a lot of instruction data where we have images, questions, corresponding answers. And we really leveraged a lot of the data that we had here at Brigham and MGH. We're obviously teaching hospitals. We have questions, we have existing teaching training materials and work closely with pathologists at multiple institutions to collect that data. And then finally trained a multimodal large language model where we could give it a whole slide image, start asking questions, what was in the image, and then it started generating all these entrusting morphologic descriptions. But then the challenge of course is that how do you validate this? So then we created validation data sets, validated on what multiple choice questions on free flowing questions where multiple pathologists, we had a panel of seven pathologists look through every response from our model as well as more generic models like the OpenAI, GPT-4 and BiomedCLIP and other models that are publicly available, and then compare how well this pathology specific model does in comparison to some of those other models.
(26:58):
And we found that it was very good at morphologic description.
Eric Topol (27:05):
It's striking though to think now that you have this large language model where you're basically interacting with the slide, and this is rich, but in another way, just to ask you, we talk about multimodal, but what about if you have electronic health record, the person's genome, gut microbiome, the immune status and social demographic factors, and all these layers of data, environmental exposures, and the pathology. Are we going to get to that point eventually?
Faisal Mahmood (27:45):
Yeah, absolutely. So that's what we're trying to do now. So I think that it's obviously one step at a time. There are some data types that we can very easily integrate, and we're trying to integrate those and really have PathChat as being a binder to all of that data. And pathology is a very good binder because pathology is medicine's ground truth, a lot of the fundamental decisions around diagnosis and prognosis and treatment trajectory is all sort of made in pathology. So having everything else bind around the pathology is a very good idea and indication. So for some of these data types that you just mentioned, like electronic medical records and radiology, we could very easily go that next step and build integrative models, both in terms of building the foundation model and then linking with language and getting it to generate responses and so forth. And for other data types, we might need to do some more specific training data types that we don't have enough data to build foundation models and so forth. So we're trying to expand out to other data types and see how pathology can act as a binder.
Eric Topol (28:57):
Well if anybody's going to build it, I'm betting on you and your team there, Faisal. Now what this gets us to is the point that, was it 96% or 95% of pathologists in this country are basically in an old era, we're not eking out so much information from slides that they could, and here you're kind of in another orbit, you're in another world here whereby you're coming up with information. I mean things I never thought really the prognosis of a patient over extended period of time, the sensitivity of drugs to the tumor from the slide, no less the driver mutations to be able to, so you wouldn't even have to necessarily send for mutations of the cancer because you get it from the slide. There's so much there that isn't being used. It's just to me unfathomable. Can you help me understand why the pathology community, now that I know you're not actually a pathologist, but you're actually trying to bring them along, what is the reason for this resistance? Because there's just so much information here.
Faisal Mahmood (30:16):
So there are a number of different reasons. I mean, if you go into details for why digital pathology is not actively happening. Digitizing an entire department is expensive, retaining large amounts of slides is expensive. And then the value proposition in terms of patient care is definitely there. But the financial incentives, reimbursement around AI is not quite there yet. It's slowly getting there, but it's not quite there yet. In the meantime, I think what we can really focus on, and what my group is thinking a lot about is that how can we democratize these models by using what the pathologists already have and they all have a microscope and most of them have a microscope with a camera attached to it. Can we train these models on whole slide images like we have them and adapt them to just a camera coupled to a microscope? And that's what we have done for PathChat2.
(31:23):
I think one of the demos that we showed after the article came out was that you could use PathChat on your computer with the whole slide image, but you can also use it with a microscope just coupled to a camera and you put a glass light underneath. And in an extreme lower source setting, you can also use it with just a cell phone coupled to a microscope. We're also building a lighter weight version of it that wouldn't require internet, so it would just be completely locally deployed. And then it could be active in lower source settings where sometimes sending a consult can take a really, really long time, and quite often it's not very easy for hospitals in lower source settings to track down a patient again once they've actually left because they might've traveled a long distance to get to the clinic and so forth. So the value of having PathChat deployed in a lower source setting where it can run locally without internet is just huge because it can accelerate the diagnosis so much. In particular for very simple things, which it's very, very good at making a diagnosis for those cases.
Eric Topol (32:33):
Oh, sure. And it can help bridge inequities, I mean, all sorts of things that could be an outgrowth of that. But what I still having a problem with from the work that you've done and some of the other people that well that are working assiduously in this field, if I had a biopsy, I want all the information. I don't want to just have the old, I would assume you feel the same way. We're not helping patients by not providing the information that's there just with a little help from AI. If it's going to take years for this transformation to occur, a lot of patients are going to miss out because their pathologists are not coming along.
Faisal Mahmood (33:28):
I think that one way to of course solve this would be to have it congressionally mandated like we had for electronic medical records. And there are other arguments to be made. It's been the case for a number of different hospitals have been sued for losing slides. So if you digitize all your slides and you're not going to lose them, but I think it will take time. So a lot of hospitals are making these large investments, including here at the Brigham and MGH, but it will take time for all the scanners, all the storage solutions, everything to be in place, and then it will also take time for pathologists to adapt. So a lot of pathologists are very excited about the new technology, but there are also a lot of pathologists who feel that their entire career has been diagnosing cases or using a microscope and slide. So it's too big of a transition for them. So I think there'll obviously be some transition period where both would coexist and that's happening at a lot of different institutions.
Eric Topol (34:44):
Yeah, I get what you're saying, Faisal, but when I wrote Deep Medicine and I was studying what was the pathology uptake then of deep learning, it was about 2% and now it's five years later and it's 4% or 5% or whatever. This is a glacial type evolution. This is not keeping up with how the progress that's been made. Now, the other thing I just want to ask you before finishing up, there are some AI pathology companies like PathAI. I think you have a startup model Modella AI, but what can the companies do when there's just so much reluctance to go into the digital era of pathology?
Faisal Mahmood (35:31):
So I think that this has been a big barrier for most pathology startups because around seven to eight years ago when most of these companies started, the hope was that digital pathology would happen much faster than it actually has. So I think one thing that we're doing at Modella is that we understand that the adoption of digital pathology is slow. So everything that we are building, we're trying to enable it to work with the current solutions that exist. So a pathologist can capture images from a pathology slide right in their office with a camera with a microscope and PathChat, for example, works with that. And then the next series of tools that we're developing around generative AI would also be developed in a manner that it would be possible to use just a camera coupled to a microscope. So I think that I do feel that all of these pathology AI companies would have been doing much, much better if everything was digital, because adopting the tools that they developed would very straightforward. Right now, the barrier is that even if you want to deploy an AI driven solution, if your hospital is not entirely digital, it's not possible to do that. So it requires this huge upfront investment.
Eric Topol (37:06):
Yeah, no, it's extraordinary to me. This is such an exciting time and it's just not getting actualized like it could. Now, if somebody who's listening to our conversation has a relative or even a patient or whatever that has a biopsy and would like to get an enlightened interpretation with all the things that could be found that are not being detected, is there a way to send that to a center that is facile with this? Or if that's a no go right now?
Faisal Mahmood (37:51):
So I think at the moment it's not possible. And the reason is that a lot of the generic AI tools are not ready for this. The models are very, very specific for specific purposes. The generalist models are just getting started, but I think that in the years to come, this would be a competitive edge for institutions who do adopt AI. They would definitely have a competitive edge over those who do not. We do from time to time, receive requests from patients who want us to run their slides on the cancers of unknown primary tool that we built. And it depends on whether we are allowed to do so or not, because it has to go through a regular diagnostic first and how much information can we get from the patient? But it's on a case by case basis.
Eric Topol (38:52):
Well, I hope that's going to change soon because you have been, your team there has just been working so hard to eke out all that we can learn from a path slide, and it's extraordinary. And it made me think about what we knew five years ago, which already was exciting, and you've taken that to the fifth power now or whatever. So anyway, just to congratulate you for your efforts, I just hope that it will get translated Faisal. I'm very frustrated to learn how little this is being adopted here in this country, a rich country, which is ignoring the benefits that it could provide for patients.
Faisal Mahmood (39:40):
Yeah. That's our goal over the next five years. So the hope really is to take everything that we have developed so far and then get it in aligned with where the technology currently is, and then eventually deploy it both at our institution and then across the country. So we're working hard to do that.
Eric Topol (40:03):
Well, maybe patients and consumers can get active about this and demand their medical centers to go digital instead of living in an analog glass slide world, right? Yeah, maybe that's the route. Anyway, thank you so much for reviewing at this pace of your publications. It's pretty much unparalleled, not just in pathology AI, but in many parts of life science. So kudos to you, Richard Chen, and your group and so many others that have been working so hard to enlighten us. So thanks. I'll be checking in with you again on whatever the next model that you build, because I know it will be another really important contribution.
Faisal Mahmood (40:49):
Thank you so much, Eric. Thanks.
**************************
Thanks for listening, reading or watching!
The Ground Truths newsletters and podcasts are all free, open-access, without ads.
Please share this post/podcast with your friends and network if you found it informative
Voluntary paid subscriptions all go to support Scripps Research. Many thanks for that—they greatly helped fund our summer internship programs for 2023 and 2024.
Thanks to my producer Jessica Nguyen and Sinjun Balabanoff for audio and video support at Scripps Research.
Side note: My X/twitter account @erictopol was hacked yesterday, 27 July, with no help from the platform to regain access despite many attempts. Please don’t get scammed!











Faisal Mahmood: A.I.'s Transformation of Pathology