The First Diagnostic Immunome
A Big Step Forward By Sequencing B and T Cell Receptors Plus A.I.
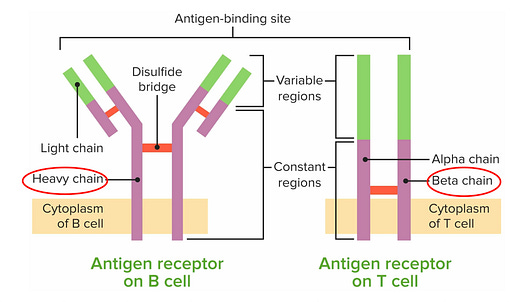
B and T lymphocytes contain a treasure chest of information about our immune response to pathogens, vaccination, environmental exposures, and untoward, self-directed attack (to our organs, tissues, and cells). The B and T cell receptors are like antennas, extending outside the cell (Figure below), that act as the sensors to mount a response to these different antigens. Their response is stored, but these receptors are exceedingly complex to decode because their diversity is extreme, with extensive gene rearrangements and somatic mutations. Until now, sequencing these receptors has not been used to diagnose an autoimmune condition or exposure to a pathogen. Their only clinical use case to date has been quite rare, in certain lymphomas for diagnosis and management.
Many autoimmune diseases are hard to accurately diagnose, require panels of expensive tests, and often lead to substantial delays. In fact, the average patient with an autoimmune disease requires visits with 4 doctors over 4.5 years to get an accurate diagnosis and treatment plan. The field has been waiting for a demonstration to unlock the potential of B and T cell repertoires—the complete collection of receptors express within an individual. In my prior post on Ground Truths, I outlined why it is so essential to have an immunome, since the only routine lab test we have today to assess a person’s immune system is the ratio of neutrophils to lymphocytes, a crude marker, what can be considered as an icepick view of the body’s most complex system. I’ll come back to that later, since an immunome has promise well beyond diagnosing autoimmune conditions.
“This is a one-shot sequencing approach that captures everything that your immune system has been exposed to.”—Sarah Teichmann, University of Cambridge
Enter Mal-ID (Machine Learning for Immunological Diagnosis)
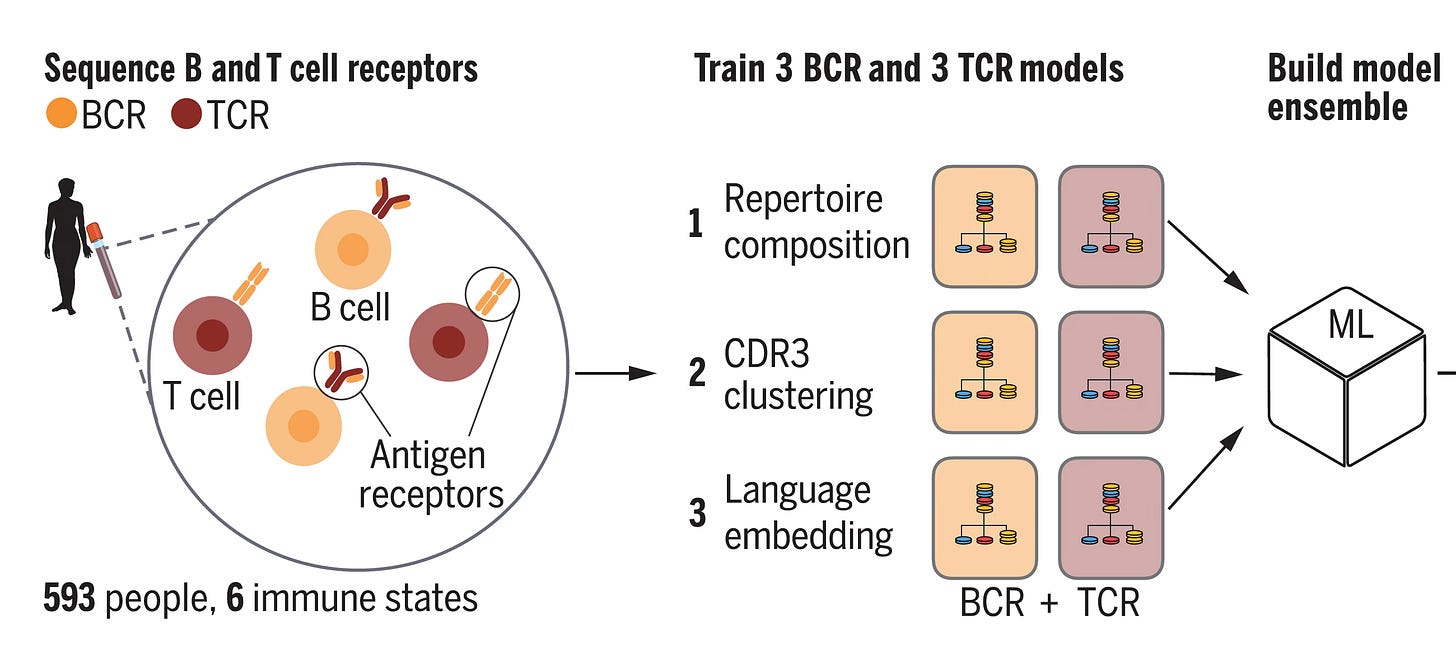
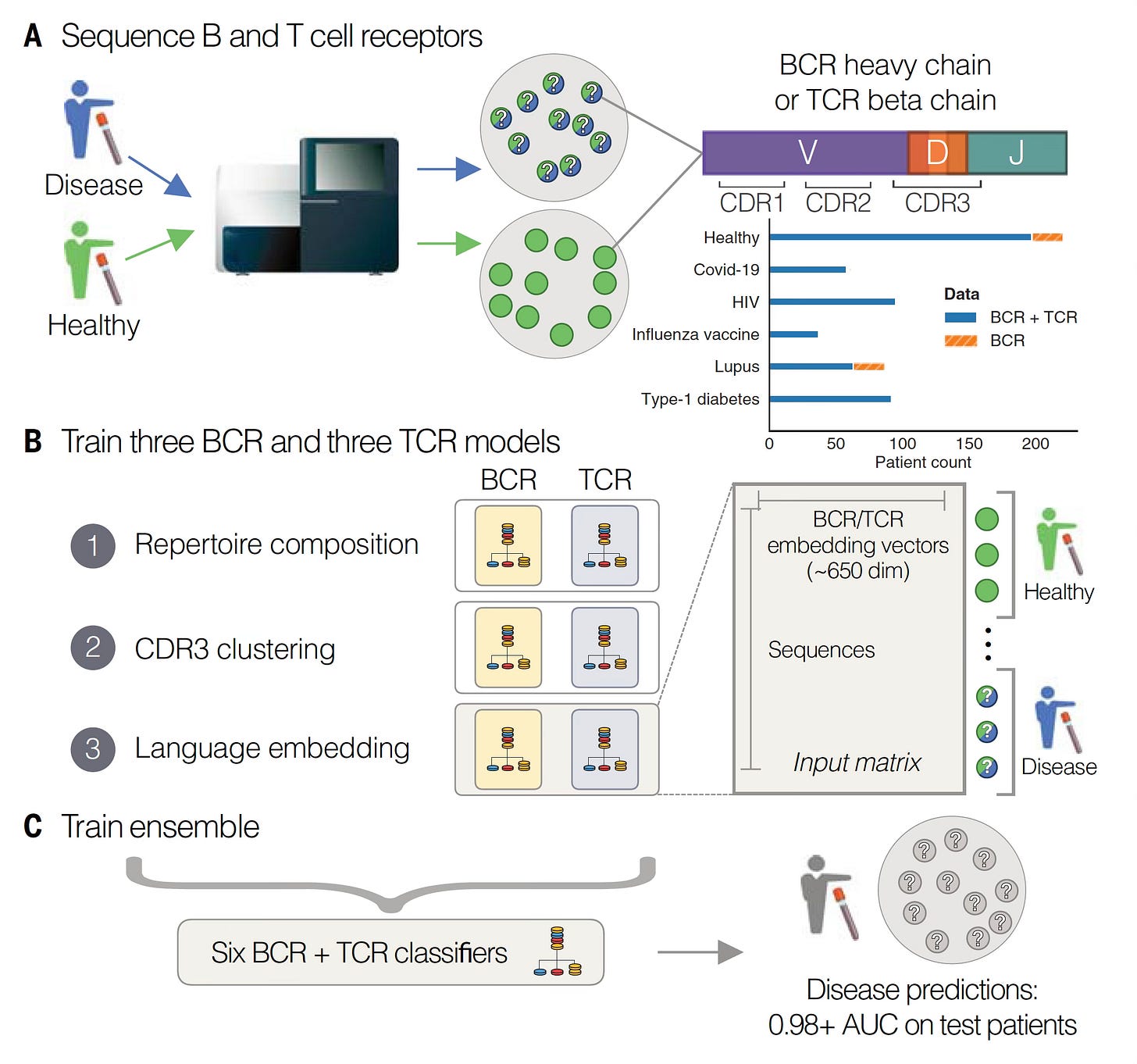
In Science this week, Maxim Zaslavsky and colleagues (and a who’s who list of leading immunologists and rheumatologists in the US and Europe) published an extraordinary paper, the first sequencing of B (BCR) and T cell receptors (TCR) at scale across multiple diagnoses. This work included 63 people with acute Covid who were hospitalized, 86 with lupus (some were pediatric cases), 92 with autoimmune Type 1 diabetes, 95 with HIV, 37 who had a recent flu shot, and 220 different controls. A total of nearly 600 individuals (N=593). From blood samples, they sequenced the heavy chain of the B cell receptor and the beta chain of the T cell receptor (as seen in the Figure above) in tens of millions of lymphocytes.
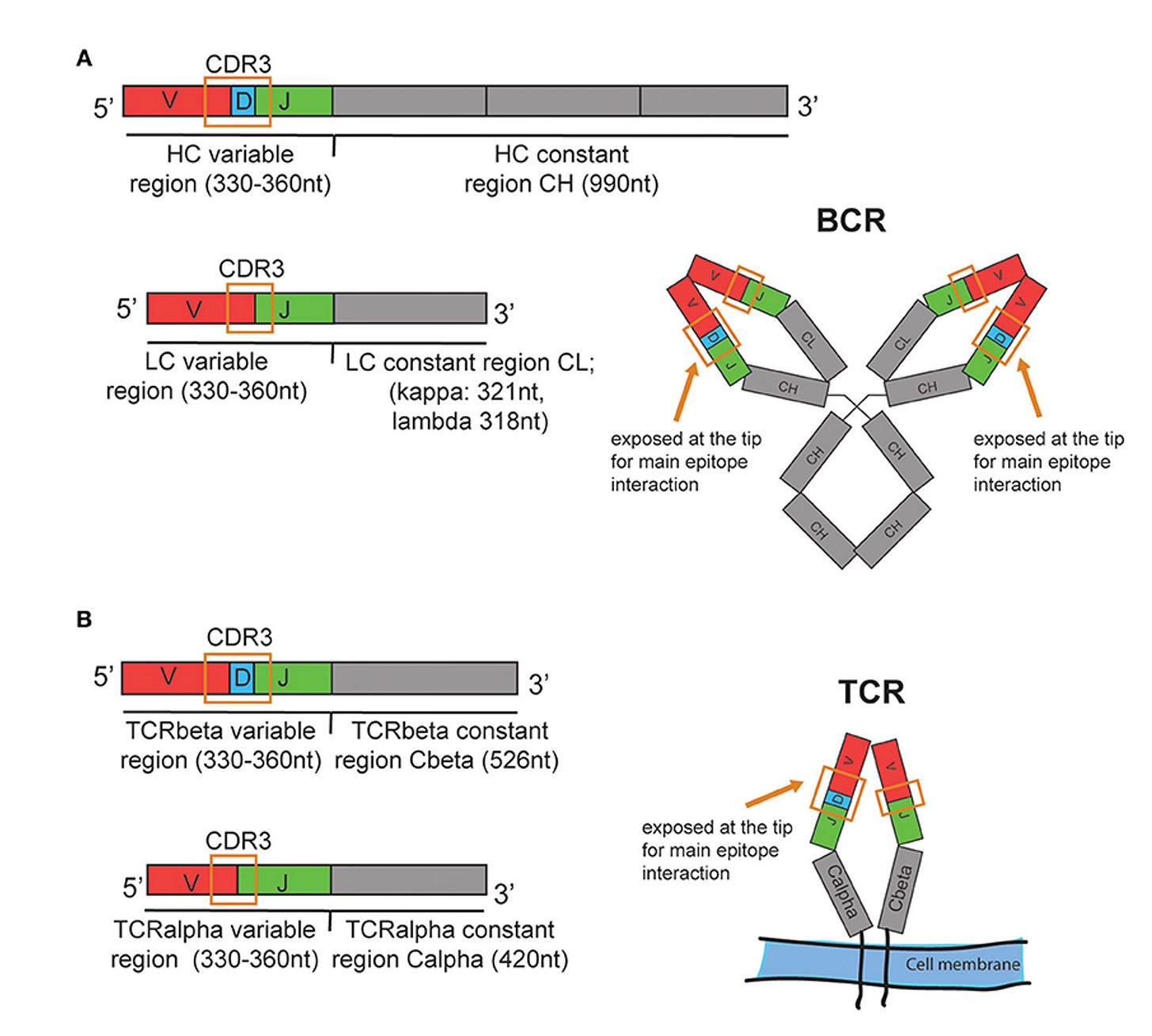
Before we get into their machine learning and A.I. models, it’s helpful to review a bit more about these lymphocyte receptors (Figure below). Their business ends, at the tips, are the variable regions which contain 3 different genomic segments: V for variable, D for diversity and J for joining. There are complementary-determining regions (CDRs) that are key loops in the receptors that determine binding to the antigens. Note CDR3, which has the highest specificity for recognizing and binding an antigen, which can be considered a unique fingerprint of each receptor.
Three Models for Each Receptor and an Ensemble for All 6
With that brief grounding on structure and function of these receptors, let’s review the 3 different models used for analysis of the data, to find the key patterns in a massive dataset. To emphasize, this is an example of agnostic analytics—the BCR and TCR sequence data are inputs to the models without any annotation or connection with clinical of health conditions (or anything else for that matter). As seen below, Model 1, called Repertoire Composition, had inputs of the frequency of different gene segments. Model 2 focused on CDR3, assessing the sequences and clusters of these sequences in individuals with the same condition. Both Model 1 and 2 were straightforward machine learning or logistic regression type analytics. Model 3 was different, since it used an open-source, large language model called Evolutionary Scale Model-2 (ESM-2) for proteins, a type of generative A.I. based on transformer architecture that is self-supervised, not requiring any training for input of these data (which was CDR3 based). The 3 models were each applied to the dataset for BCR and TCR and all 6 were combined into the Ensemble model. (NB: this is an oversimplified description of complex models, see the Methods section of the paper for details.)
Note that some individuals did not have both BCR and TCR sequence data (top right in digram below). Complete data were available in 542 individuals. Some of the individuals had multiple samples.
The Main Results
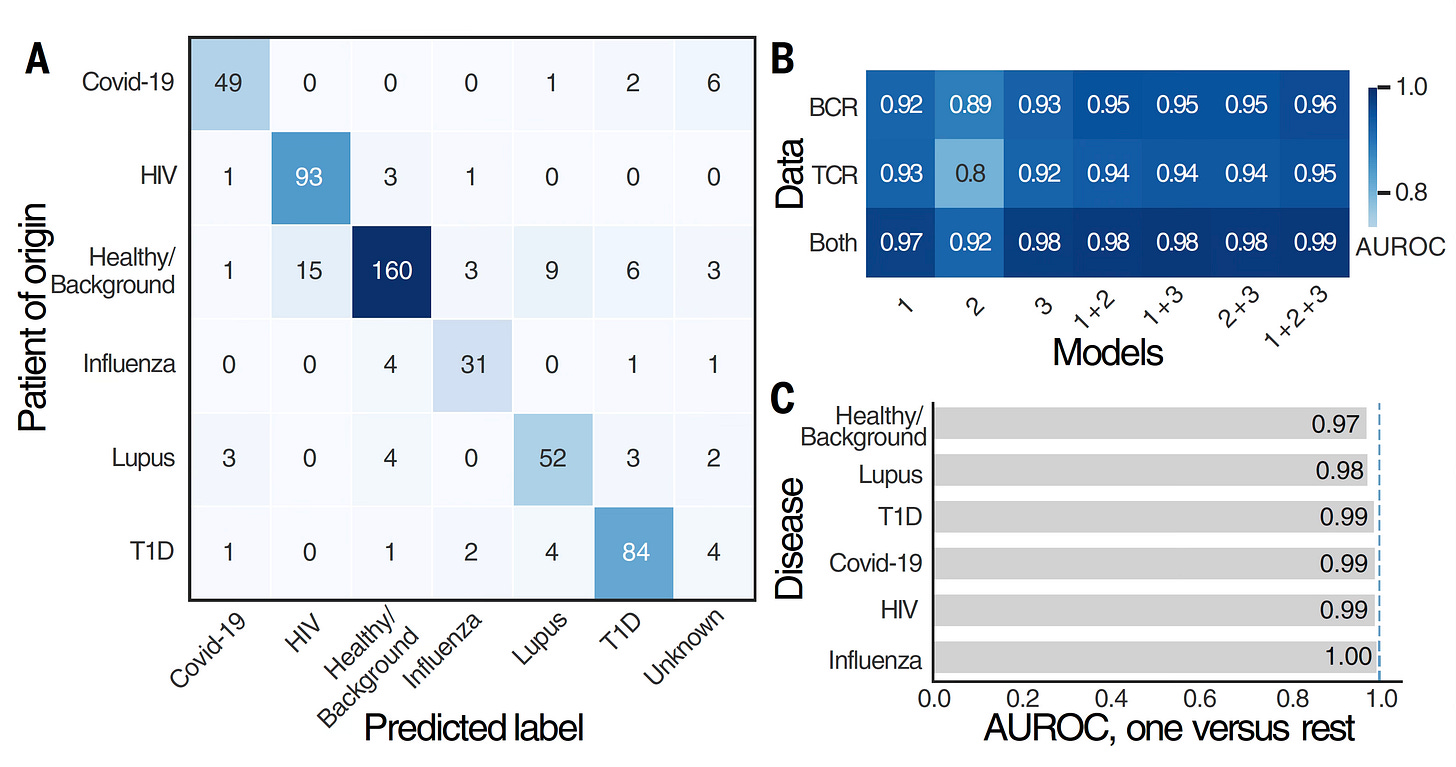
Below are the classification by BCR and TCR for the different diagnoses, with remarkable accuracy, an area under the receiver operative characteristic curve (AUROC) of 0.986 (1.0 = perfect). It’s a measure of model performance reflecting accuracy for correct classification. Usually when you’d see such a high value, it would be considered too good to be true. I’ll get to the reason for such a high AUROC a little later. You’ll note that having both BCR and TCR data brought the level of accuracy higher, maxed out with all 3 models (1+2+3) at 0.99 (top right). Although it might seem an increase in AUROC from 0.93 to 0.98 (Model 3 alone) is not much numerically, it actually is meaningful as can be distinguished in the confusion matrix displays of data (one of which is in Panel A below, many in the extensive Supplemental data).
That’s different from diagnostic specificity and sensitivity. For lupus it was 93% and 90%, respectively. Overall, for individuals in the disease categories, the models classified 93% of individuals accurately (as not healthy) and 87.5% were categorized correctly specific to their disease.
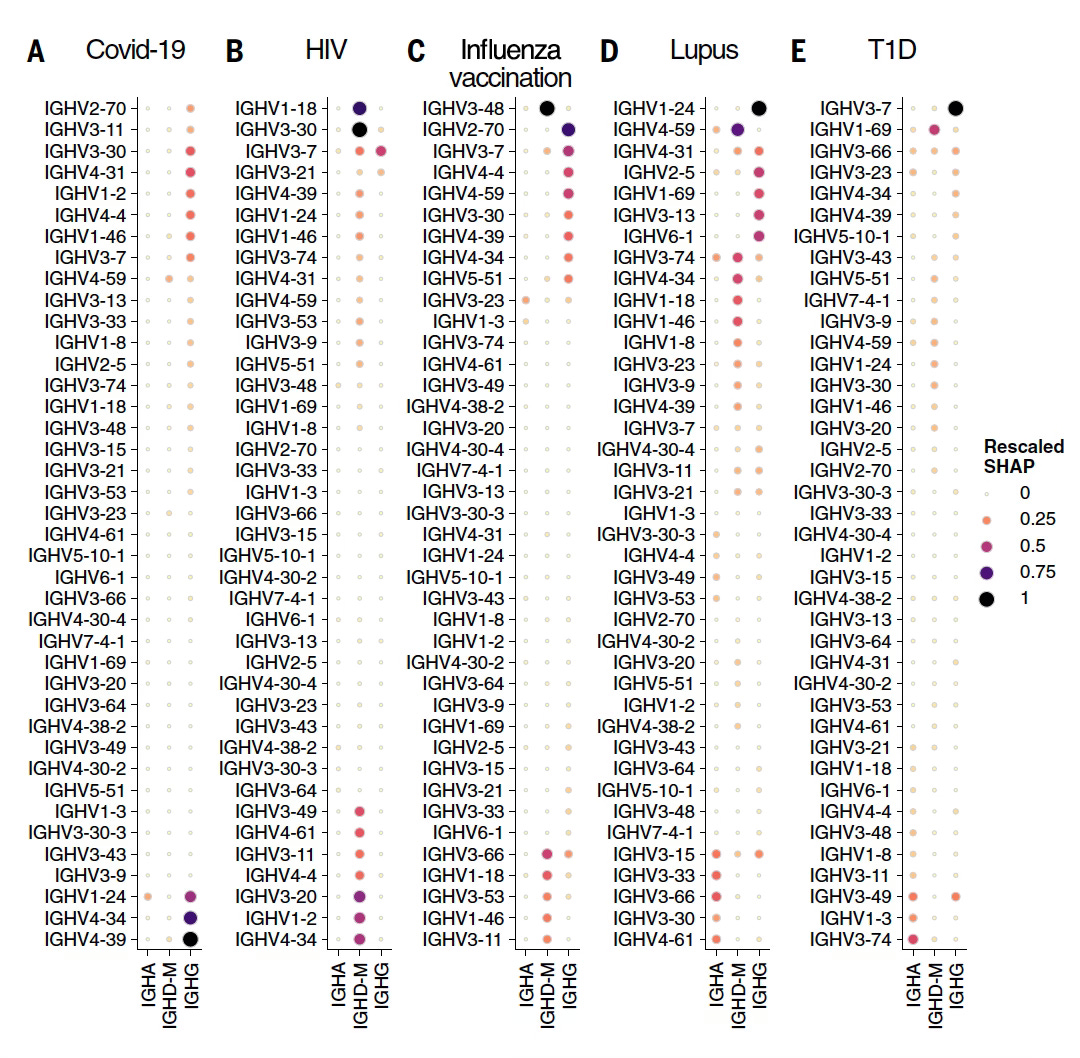
The other main output of results pertained to identifying specific patterns in the BCR and TCR genes related to particular conditions (output from Model 3 below, SHAP=Shapley importance of ranking). These data were presented as immunoglobulin heavy chain variable region (IGHV) for each condition, and were decomposed to isotopes (specific antibodies) such as in IgA or IgG. What is noteworthy is that many of these genes correlated with known underpinnings of the various conditions, such as lupus, type 1 diabetes, or HIV, and several were new loci that may turn out to expand upon our current knowledge base.
Some Critique of the Analysis
“This is the first study with a sufficient number of donors, a sufficient depth of sampling of each of those donors, a significant enough distribution of disease states, and then a sophisticated enough modeling that they were able to actually make this work”—Bryan Briney, Scripps Research
Very Good Features included:
—Separate training and validation: “We ensured that models were never trained on data from a patient and then evaluated on other data from the same person.
—Extensive cross-validation, replication, external validation, sample protocols from different labs
—Tests for age, sex and ethnicity showed limited impact
—Tests for batch effects showed limited impact
—Showing incremental information compared with clinical data from Mal-ID.
—Showing how the 3 models together provided additivity for accuracy of categorization, supporting their complementary informativeness
Some Limitations included:
—Not a prospective assessment
—A multiple choice, constrained classification which boosted model performance (AUROC)
—The number of diseases assessed were limited for a few autoimmune conditions; people can have more than one; the model is only as good as what it is trained on
—Some healthy people were misclassified, the issue of false positives—but note it may not actually be a misclassification…it could represent vulnerability to an autoimmune condition (think non-binary)
—Unknown level of accuracy, predictive value for what would lead to clinical acceptance (what should be cutoffs?)
—Single point of TCR and BCR assessment ; longitudinal sampling should be more informative
Significance and Contextualization
Yesterday evening, I had a great opportunity to have a 1-hour zoom session with the principal authors of the paper, Maxim Zaslavsky, Anshul Kundaje, and Scott Boyd (who I had never met or known) to discuss the paper and their plans going forward. Since the work was published, they have done BCR and TCR sequencing in many more people with autoimmune conditions and are also seeing a signal for response to therapy in these patients. It was clear from our discussion that BCR and TCR could be done at scale, inexpensively, in batches with a couple of days turnaround. While they haven’t formed a start-up company, they, along with some of the other authors, have filed patent applications.
These are my takeaways from the paper, our conversation, and what can be clincally gleaned from an immunome.
This work could not have been accomplished without A.I., specifically advanced with their use of a large language model (ESM-2). The complexity of BCR and TCR datasets are especially well suited for (indeed require) GenAI analytics.
“So it’s a very complicated challenge, and maybe one that is ripe for AI and large language models to jump into.”—Marion Pepper, University of Washington
The paper provides a first proof-of-concept, a stepping stone, towards a full immunome. The experiment can be considered as “contrived” since there were a very limited number of conditions assessed on a retrospective basis. Nevertheless, the results can be viewed as remarkable, a first-shot diagnostic for some autoimmune conditions. My discussion with the authors pointed to much more progress, expansion taking place since the publication.
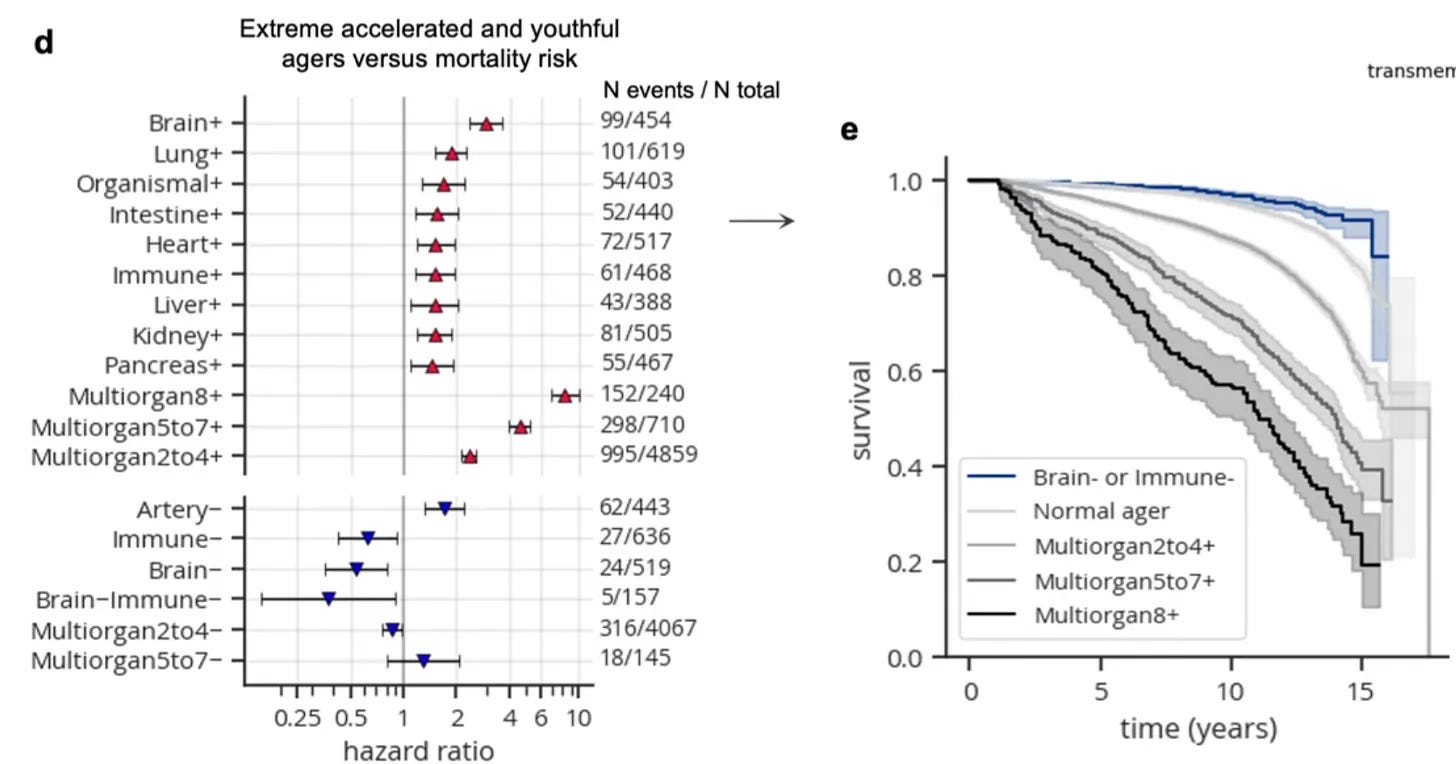
Ultimately, a full immunome can be “panoramic,” BCR and TCR sequencing Integrated with other immune system components (autoantibodies, virus and pathogen exposure, flow cytometry, T cell functional assays, interferon, HLA typing, etc). For autoimmune conditions, this (BCR and TCR sequencing) could add to current rheumatological panels that include rheumatoid factor, double-stranded DNA, anti-nuclear antibody, anti-cyclic citrullinated peptide antibodies, and others. Combined with this would be proteomic organ clocks, such as what I’ve previously reviewed for the immune system aging clock, prominently linked with healthspan and life expectancy. (Figure below is from that prior post.)
The clinical use cases for BCR and TCR are exceptionally broad. We obviously do not need these to diagnose Covid or flu. But diagnosing and following individuals with Long Covid is an exemplar for how this could be applied, testing the long list of candidate interventions that may suppress the autoimmune features of this complex condition. Not just diagnosing but preventing autoimmune conditions (well before there has been any tissue destruction such as in multiple sclerosis or lupus) in people at high-risk looms as potential ways to apply insights from B and T cell sequencing.
In the future, an immunome will become a routine assessment for medicine, which I lay out in my new book SUPER AGERS, that will be published in a couple of months. Our immune system is inextricably linked to the major age-related diseases—cardiovascular, cancer and neurodegenerative. Until now we haven’t had a way to assess its status, which we know exhibits marked inter-individual variability with aging. We now have far better ways to control—rev up or suppress— our immune system than ever before (and more are emerging quickly). That’s happening at the same time our capability for precision medical forecasting is blossoming, using multimodal A.I. for integration of all of a person’s data to determine whether they are at any high-risk for a major age-related condition or acceleration of organ aging (such as brain, heart, kidney, immune system) and when.
******************************************************************
Thanks for reading and subscribing to Ground Truths.
If you found this interesting please share it!
That makes the work involved in putting these together especially worthwhile.
All content on Ground Truths—its newsletters, analyses, and podcasts, are free, open-access.
Paid subscriptions are voluntary and all proceeds from them go to support Scripps Research. They do allow for posting comments and questions, which I do my best to respond to. Please don't hesitate to post comments and give me feedback. Many thanks to those who have contributed—they have greatly helped fund our summer internship programs for the past two years.
N.B. These are dark times for biomedicine in the United States, with the changes that are occurring at HHS, NIH, FDA, and the CDC. Ironically, this is occurring at a time when there is a marked acceleration of progress, some of which is being propelled by genAI as highlighted in this post and previous ones (such as language of life models and high-throughput proteomics). I haven’t lost my optimism. I think about how eventually Newton’s Third Law (for every action, there is an equal and opposite reaction) will come into play and the pendulum will swing backward—what I really mean is forward!


I'm coming at this from a functional medicine perspective, which understands for starters that a medical "condition" can have a variety of underlying causes, and that different conditions will sometimes have the same underlying cause.
Thus, if you're looking for a root-cause solution to a set of symptoms, it's actually the underlying causes that you need to elucidate.
We believe root cause solutions are preferable where available.
This type of tool could go in that direction, assuming specific B and T cell derangements can be linked to specific underlying causes.
Otherwise, we're stuck throwing inhibitors of this or that pathway or cytokine at people, perhaps with more success than we have had to date, but I don't think that's assured.
Hope these methods can also be done for ME/cfs that is disabling millions, and similar to Long Covid, Chronic Lyme etc. If these researchers could collaborate with SolveME.org, OMF.NGO, Polybio.org, Bateman Horne Center and others that see many clients, this could help millions in US alone.