David Liu is an gifted molecular biologist and chemist who has pioneered major refinements in how we are and will be doing genome editing in the future, validating the methods in multiple experimental models, and establishing multiple companies to accelerate their progress.
The interview that follows here highlights why those refinements beyond the CRISPR Cas9 nuclease (used for sickle cell disease) are vital, how we can achieve better delivery of editing packages into cells, ethical dilemmas, and a future of somatic (body) cell genome editing that is in some ways is up to our imagination, because of its breadth, over the many years ahead.
Recorded 29 November 2023 (knowing the FDA approval for sickle cell disease was imminent)
Annotated with figures, external links to promote understanding, highlights in bold or italics, along with audio links (underlined)
Eric Topol (00:11):
Hello, this is Eric Topol with Ground Truths and I'm so thrilled to have David Liu with me today from the Broad Institute, Harvard, and an HHMI Investigator. David was here visiting at Scripps Research in the spring, gave an incredible talk which I'll put a link to. We're not going to try to go over all that stuff today, but what a time to be able to get to talk with you about what's happening, David. So welcome.
David Liu (00:36):
Thank you, and I'm honored to be here.
Eric Topol (00:39):
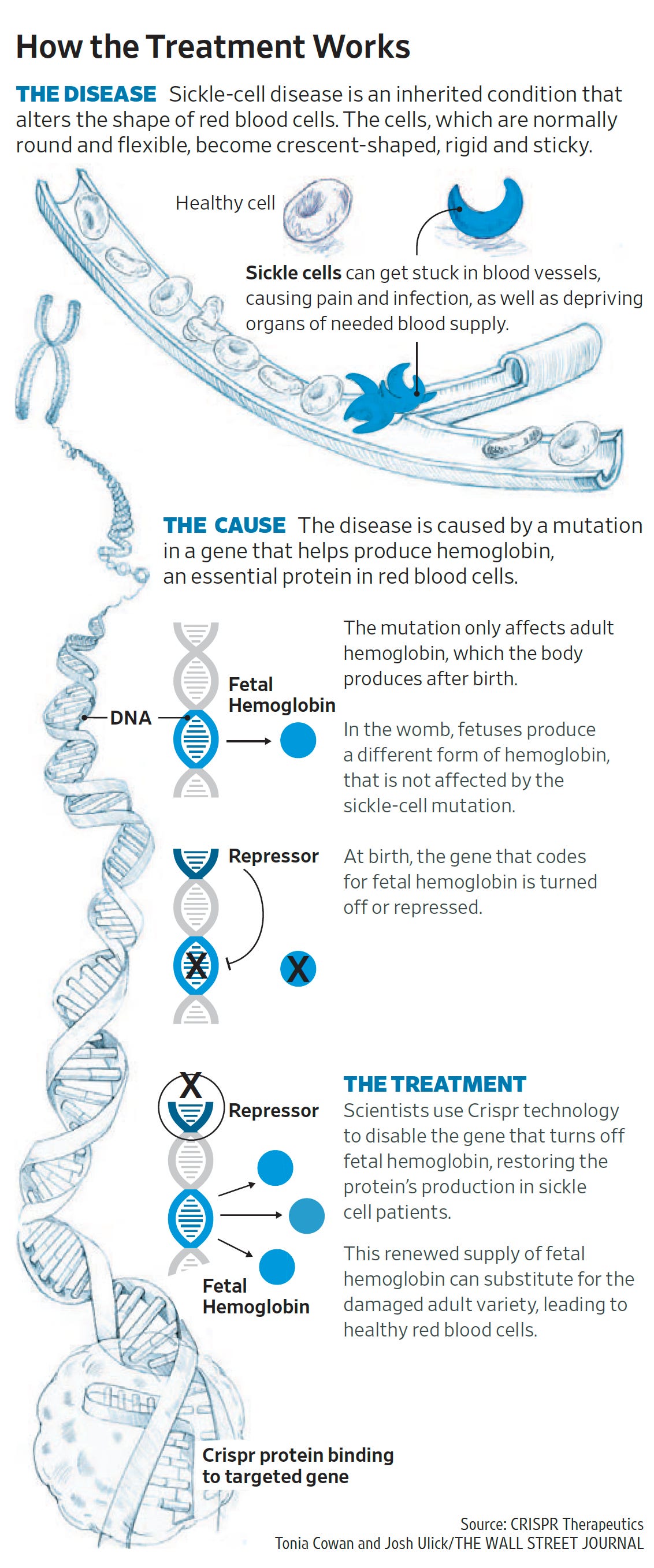
Well, the recent UK approval (November 16, 2023) of the first genome editing after all the years that you put into this, along with many other colleagues around the world, is pretty extraordinary. Maybe you can just give us a sense of that threshold that's crossed with the sickle cell and beta thalassemia also imminently [FDA approval granted for sickle-cell on 8 December 2023] likely to be getting that same approval here in the U.S.
David Liu (01:05):
Right? I mean, it is a huge moment for the field, for science, for medicine. And just to be clear and to give credit where credit is due, I had nothing to do with the discovery or development of CRISPR Cas9 as a therapeutic, which is what this initial gene editing CRISPR drug is. But of course, the field has built on the work of many scientists with respect to CRISPR Cas9, including Emmanuel Charpentier and Jennifer Doudna and George Church and Feng Zhang and many, many others. But it is, I think surprisingly rapid milestone in a long decade’s old effort to begin to take some control over our genetic features by changing DNA sequences of our choosing into sequences that we believe will offer some therapeutic benefit. So this initial drug is the CRISPR Therapeutics /Vertex drug.
Now we can say it's actually a drug approved drug, which is a Crispr Cas9 nuclease programmed to cut a DNA sequence that is involved in silencing fetal hemoglobin genes. And as you know, when you cut DNA, you primarily disrupt the sequence that you cut. And so if you disrupt the DNA sequence that is required for silencing your backup fetal hemoglobin genes, then they can reawaken and serve as a way to compensate for adult hemoglobin genes like the defective sickle cell alleles that sickle cell anemia patients have. And so that's the scientific basis of this initial drug.
Eric Topol (03:12):
So as you aptly put— frame this—this is an outgrowth of about a decade's work and it was using a somewhat constrained, rudimentary form of editing. And your work has taken this field considerably further with base and prime editing whereby you're not just making a double strand cut, you're doing nicks, and maybe you can help us understand this next phase where you have more ways you can intervene in the genome than was possible through the original Cas9 nucleases.
David Liu (03:53):
Right? So gene editing is actually a several decades old field. It just didn't quite become as popular as it is now until the discovery of CRISPR nucleases, which are just much easier to reprogram than the previous programmable zinc finger or tail nucleases, for example. So the first class of gene editing agents are all nuclease enzymes, meaning enzymes that take a piece of DNA chromosome and literally cut it breaking the DNA double helix and cutting the chromosome into two pieces. So when the cell sees that double strand DNA break, it responds by trying to get the broken ends of the chromosome back together. And we think that most of the time, maybe 90% of the time that end joining is perfect, it just regenerates the starting sequence. But if it regenerates the starting sequence perfectly and the nuclease is still around, then it can just cut the rejoin sequence again.
(04:56):
So this cycle of cutting and rejoining and cutting and rejoining continues over and over until the rejoining makes the mistake that changes the DNA sequence at the cut site because when those mistakes accumulate to a point that the nuclease no longer recognizes the altered sequence, then it's a dead end product. That's how you end up with these disrupted genes that result from cutting a target DNA sequence with a nuclease like Crispr Cas9. So Crispr Cas9 and other nucleases are very useful for disrupting genes, but one of their biggest downsides is in the cells that are most relevant to medicine, to human therapy like the cells that are in your body right now, you can't really control the sequence of DNA that comes out of this process when you cut a DNA double helix inside of a human cell and allow this cutting and rejoining process to take place over and over again until you get these mistakes.
(06:03):
Those mistakes are generally mixtures of insertions and deletions that we can't control. They are usually disruptive to a gene. So that can be very useful when you're trying to disrupt the function of a gene like the genes that are involved in silencing fetal hemoglobin. But if you want to precisely fix a mutation that causes a genetic disease and convert it, for example, back into a healthy DNA sequence, that's very hard to do in a patient using DNA cutting scissors because the scissors themselves of course don't include any information that allows you to control what sequence comes out of that repair process. You can add a DNA template to this cutting process in a process called HDR or Homology Directed Repair (figure below from the Wang and Doudna 10-year Science review), and sometimes that template will end up replacing the DNA sequence around the cut site. But unfortunately, we now know that that HDR process is very inefficient in most of the types of cells that are relevant for human therapy.
(07:12):
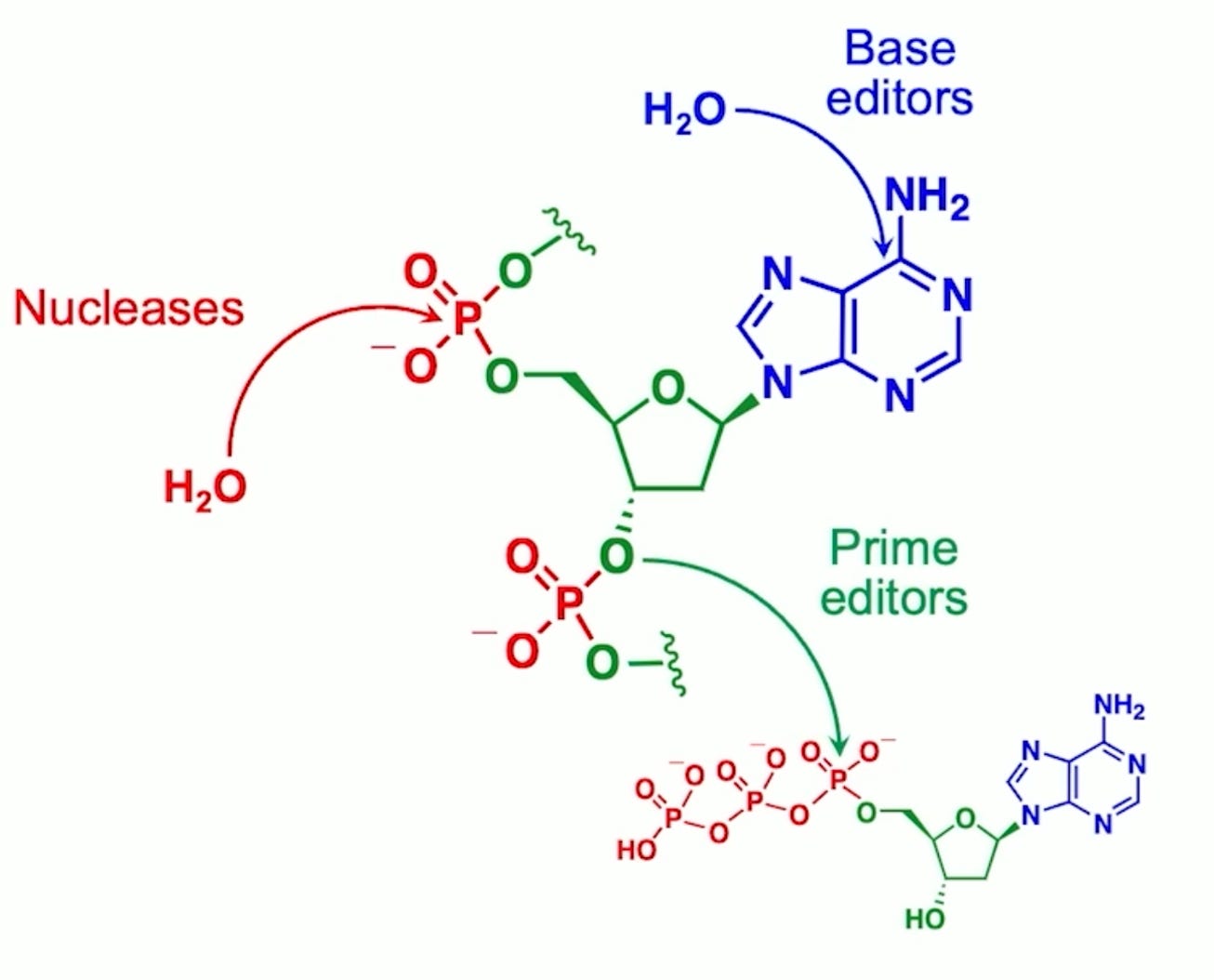
And that explains why if you look at the 50 plus nuclease gene editing clinical trials that are underway or have taken place, all but one use nucleases for gene disruption rather than for gene correction. And so that's really what inspired us to develop base editing in 2016 and then prime editing in 2019. These are methods that allow you to change a DNA sequence of your choosing into a different sequence of your choosing, where you get to specify the sequence that comes out of the editing process. And that means you can, for the first time in a general way, programmable change a DNA sequence, a mutation that causes a genetic disease, for example, into a healthy sequence back into the normal, the so-called wild type sequence, for example. So base editors work by actually performing chemistry on an individual DNA base, rearranging the atoms of that base to become a different base.
(08:22):
So base editors can efficiently and robustly change A's into G's G's, into A's T's into C's or C's into T's. Those four changes. And those four changes for interesting biochemical reasons turn out to be four of the most common ways that our DNA mutates to cause disease. So base editors can be used and have been used in animals and now in six clinical trials to treat a wide variety of diseases, high cholesterol and sickle cell disease, and T-cell leukemia for example. And then in prime editors we developed a few years later to try to address the types of changes in our genomes that caused genetic disease that can't be fixed with a base editor, for example. You can't use a base editor to efficiently and selectively change an A into a T. You can't use a base editor to perform an insertion of missing DNA letters like the three missing letters, CTT, that's the most common cause of cystic fibrosis accounting for maybe 70% of cystic fibrosis patients.
(09:42):
You can't use a base editor to insert missing DNA letters like the missing TATC. That is the most common cause of Tay-Sachs disease. So we develop prime editors as a third gene editing technology to complement nucleases and base editors. And prime editors work by yet another mechanism. They don't, again, they don't cut the DNA double helix, at least they don't cause that as the required mechanism of editing. They don't perform chemistry on an individual base. Instead, prime editors take a target DNA sequence and then write a new DNA sequence onto the end of one of the DNA strands and then sort of help the cell navigate the DNA repair processes to have that newly written DNA sequence replace the original DNA sequence. And in the process it's sort of true search and replace gene editing. So you can basically take any DNA sequence of up to now hundreds of base pairs and replace it with any other sequence of your choosing of up to hundreds of base pairs. And if you integrate prime editing with other enzymes like recombinase, you can actually perform whole gene integration of five or 10,000 base pairs, for example, this way. So prime editing's hallmark is really its versatility. And even though it's the newest of the three ways that have been robustly used to edit mammalian cells and rescue animal models of genetic disease, it is arguably the most versatile by far,
Eric Topol (11:24):
Right? Well, in fact, if you just go back to the sickle cell story as you laid out the Cas9 nuclease, that's now going into commercial approval in the UK and the US, it's more of a blunt instrument of disruption. It's indirect. It's not getting to the actual genomic defect, whereas you can do that now with these more refined tools, these new, and I think that's a very important step forward. And that is one part of some major contributions you've made. Of course, there are many. One of the things, of course, that's been a challenge in the field is delivery whereby we'd like to get this editing done in many parts of the body. And of course it's easy, perhaps I put that in quotes, easy when you're taking blood out and you're going to edit those cells and them put it back in. But when you want to edit the liver or the heart or the brain, it gets more challenging. Now, you did touch on one recent report, and this is of course the people with severe familial hypercholesterolemia. The carriers that have LDL cholesterol several hundred and often don't respond to even everything we have on the shelf today. And there were 10 people with this condition that was reported just a few weeks ago. So that's a big step forward.
David Liu (13:09):
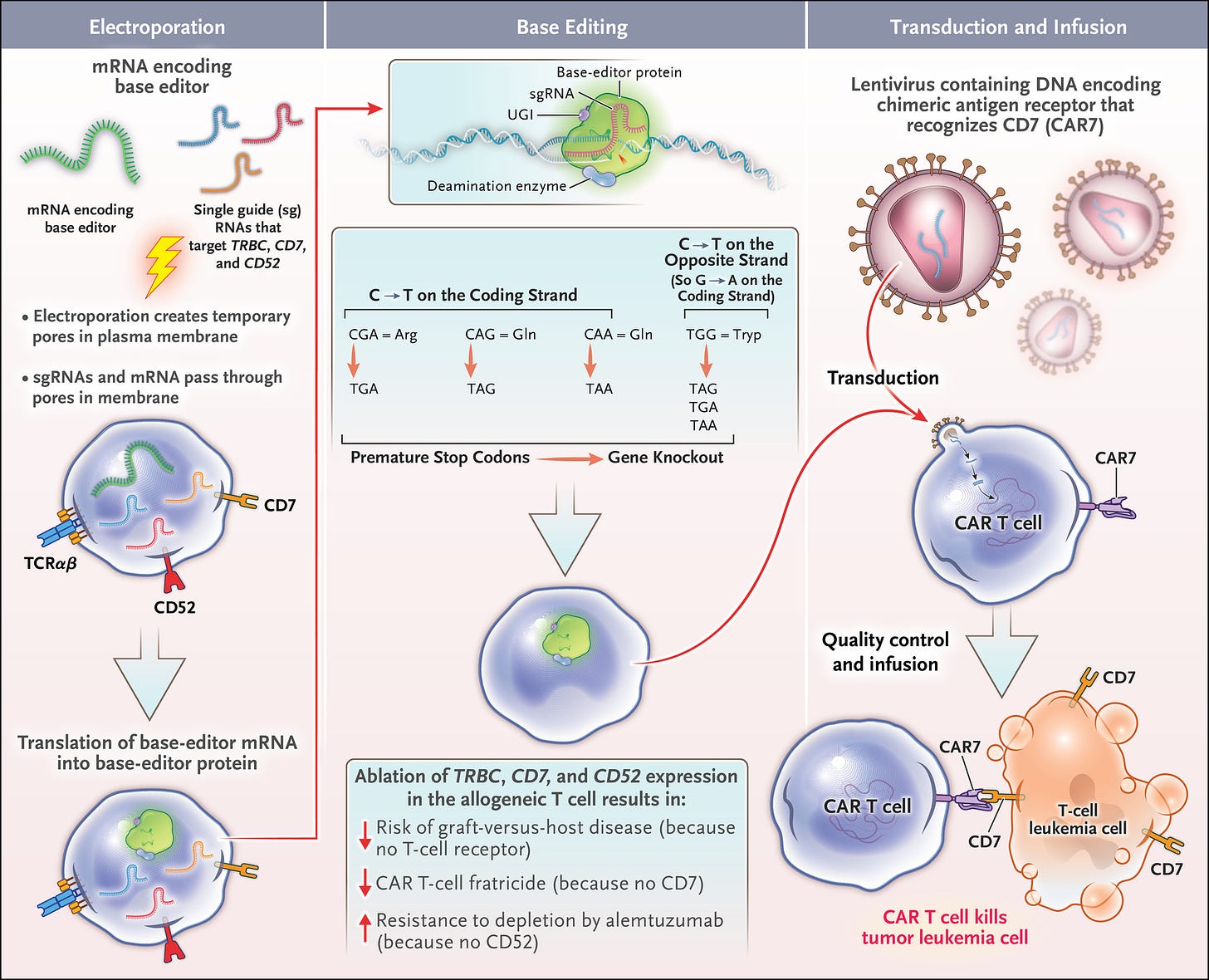
That was also a very exciting milestone. So that clinical trial was led by scientists at Verve Therapeutics and Beam Therapeutics, and it was the first clinical readout of an in vivo base editing clinical trial. There was previously at the end of 2022, the first clinical readout of an ex vivo base editing clinical trial using CAR T cells, ex vivo base edited to treat T-cell leukemia in pediatric patients in the UK. Ffigure from that NEJM paper below). But as you point out, there are only a small fraction of the full range of diseases that we'd like to treat with gene editing and the types of cells we'd like to edit that can be edited outside of the body and then transplanted back into the body. So-called ex vivo editing. Basically, you can do this with cells of some kind of blood lineage, hematopoietic stem cells, T-cells, and really not much else in terms of editing outside the body and then putting back into the body as you point out.
(14:17):
No one's going to do that with the brain or the heart anytime soon. So what was very exciting about the Verve Beam clinical trial is that Verve sought to disrupt the function of PCSK9 storied, gene validated by human genetics, because there are humans that naturally have mutations in PCSK9, and they tend to have much lower incidences of heart disease because their LDL, so-called bad cholesterol, is much lower than it would otherwise be without those mutations. So Verve set out to simply disrupt PCSK9 through gene editing. They didn't care whether they used a nuclease or a base editor. So they compared side-by-side the results of disrupting PCSK9 with Cas9 nuclease versus disrupting it by installing a precise single letter base edit using an adenine base editor. And they actually concluded that the base editor gave them higher efficacy and fewer unwanted consequences.
(15:28):
And so they went with the base editor. So the clinical trial that just read out were patients treated in New Zealand, in which they were given a lipid nanoparticle mRNA complex of an adenine base editor programmed with a guide RNA to install a specific A to G mutation in a splice site in PCSK9 that inactivates the gene so that it can no longer make functional PCSK9 protein. And the exciting result that read out was that in patients that receive this base editor, a single intravenous injection of the base editor lipid nanoparticle complex, as you know, lipid nanoparticles very efficiently go to the liver. In most cases, PCSK9 was edited in the liver and the result was substantial reduction in LDL cholesterol levels in these patients. And the hope and the anticipation is that that one-time treatment should be durable, should be more or less permanent in these patients. And I think while the patients who are at highest risk of coronary artery disease because of their genetics that give them absurdly high LDL cholesterol levels, that makes the most sense to go after those patients first because they are at extremely high risk of heart attacks and strokes. If the treatment proves to be efficacious and safe, then I think it's tempting to speculate that a larger and larger population of people who would benefit from having lower LDL cholesterol levels, which is probably most people, that they would also be candidates for this kind of therapy.
Eric Topol (17:22):
Yeah, no, it's actually pretty striking how that could be achieved. And I know in the primates that were done prior to the people in New Zealand, there was a very durable effect that went on well over I think a year or even two years. So yeah, that's right. Really promising. So now that gets us to a couple of things. One of them is the potential for off-target effects. As you've gotten more and more with these tools to be so precise, is the concern that you could have off-target effects just completely, of course inadvertent, but potential for other downstream in time known unknowns, if you will. What are your thoughts about that?
David Liu (18:15):
Yeah, I have many thoughts on this issue. It's very important the FDA and regulatory bodies are right to be very conservative about off-target editing because we anticipate those off targets will be permanent, those off-target edits will be permanent. And so we definitely have a responsibility to minimize adding to the mutational burden that all humans have as a function of existing on this planet, eating what we eat, being bombarded by cosmic rays and sunlight and everything else. But I think it's also important to put off-target editing into some context. One context is I think virtually every substance we've ever put into a person, including just about every medicine we've ever put into a person, has off-target effects, meaning modulates the function of biological molecules other than the intended target. Of course, the stakes are higher when those are gene editing agents because those modifications can be permanent.
(19:18):
I think most off-target edits are very likely to have no consequence because most of our genome, if you mutate in the kinds of small ways like making an individual base pair change for a base editor are likely to have no consequence. We sort of already know this because we can measure the mutational burden that we all face as a function of living and it's measurable, it's low, but measurable. I've read some papers that estimate that of the roughly 27 trillion [should be ~37] cells in an adult person, that there are billions and possibly hundreds of billions of mutations that accumulate every day in those 27 [37] trillion cells. So our genomes are not quite the static vaults that we'd like to think that they are. And of course, we have already purposefully given life extending medicines to patients that work primarily by randomly mutating their genomes. These are chemotherapeutic agents that we give to cancer patients.
(20:24):
So I think that history of giving chemotherapeutic agents, even though we know those agents will mess up the genomes of these patients and potentially cause cancer far later down the road, demonstrates that there are risk benefit situations where the calculus favors treatment, even if you know you are causing mutations in the genome, if the condition that the patient faces and their prognosis is sufficiently grave. All that said, as I mentioned, we don't want to add to the mutational burden of these patients in any clinically relevant way. So I think it is appropriate that the early gene editing clinical candidates that are in trials or approved now are undergoing lots and lots of scrutiny. Of course, doing an off-target analysis in an animal is of limited value because the animal's genome is quite different than the human genome. So the off targets won't align, but doing off-target analysis in human cells and then following up these patients for a long time to confirm hopefully that there isn't clinical evidence of quality of life or lifespan deterioration caused by off-target editing, that's all very, very important.
(21:55):
I also think that people may not fully appreciate that on target editing consequences also need to be examined and arguably examined with even more urgency than off-target edits. Because when you are cutting a chromosome at a target site with a nucleus, for example, you generate a complex mixture of different products of different DNA sequences that come out, and the more sequences you sequence, the more different products you realize are generated. And I don't think it's become routine to try to force the companies, the clinical groups that are running these trials to characterize the top 1000 on target products for their biological consequence. That would be sort of impractical to do and would probably slow down greatly the benefit of these early nuclease clinical trials for patients. But those are actually the products that are generated with much higher frequency typically than the off-target edits. And that's part of why I think it makes more sense from a clinical safety perspective to use more precise gene editing methods like base editing and prime editing where we know the products that are generated are mostly the products that we want are not uncontrolled mixtures of different deletion and insertion products.
(23:27):
So I think paying special attention to the on-target products, which are generated typically 70 to 100% of the time as opposed to the off targets which may be generated at a 0.1 to 1% level and usually not that many at that level once it reaches a clinical candidate. I think that's all important to do.
Eric Topol (23:51):
You've made a lot of great points there and thanks for putting that in perspective. Well, let's go on to the delivery issue. You mentioned nanoparticles, viral vectors, and then you've come up with small virus-like neutered viruses if you will. I think a company Nvelop that you've created to push on that potential. What are your thoughts about where we stand since you've become a force for coming up with much better editing, how about much better and more diverse delivery throughout the body? What are your thoughts about that?
David Liu (24:37):
Yeah, great. Great question. I think one of the legacies of gene editing is and will be that it inspired many more scientists to work hard on macromolecular delivery technologies. All of these gene editing agents are macromolecules, meaning they’re proteins and or nucleic acids. None of them are small molecules that you can just pop a pill and swallow. So they all require special technologies to transfer the gene editing agent from outside of the cell into the cell. And the fact that taking control of our genetic features has become such a popular aspiration of medicine means that there's a lot of scientists as measured, most importantly by the young scientists, by the graduate students and the postdocs and the young professors of which I'm no longer one sadly, who have decided that they're going to devote a big part of their program to delivery. So you summarized many of the clinically relevant, clinically validated delivery technologies already, somewhat sadly, because if there were a hundred of these technologies, you probably wouldn't need to ask this question. But we have lipid nanoparticles that are particularly good at delivering messenger RNA, that was used to deliver the covid vaccine into billions of people. Now also used to deliver, for example, the adenine base editor mRNA into the livers of those hypercholesterolemia patients in the Verve/Beam clinical trial.
(26:20):
So those lipid nanoparticles are very well matched for gene editing delivery as long as it's liver. And they also are particularly well matched because their effect is transient. They cause a burst of gene editing agents to be produced in the liver and then they go away. The gene editing agents can't persist, they can't integrate into the genome despite what some conspiracy theorists might worry about. Not that you've had any encounter with any of those people. I'm sure that's actually what you want for a gene editing agent. You ideally want a delivery method that exposes the cell only for the shortest amount of time needed to make the on-target edit at the desired level. And then you want the gene editing agent to disappear and never come back because it shouldn't need to. DNA edits to our genome for durable cells should be permanent. So that's one method
.
(27:25):
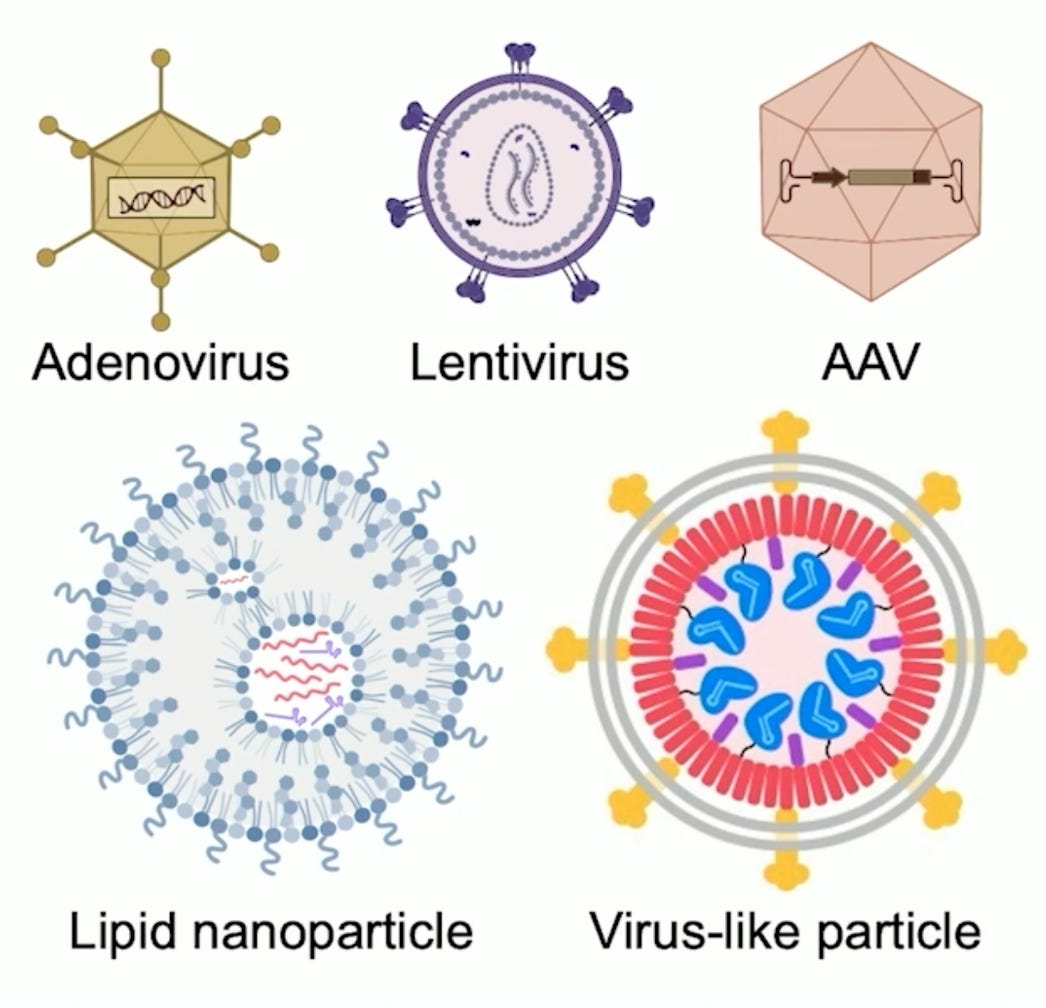
And then there are a variety of other methods that researchers have used to deliver to other cells, but they each carry some trade-offs. So if you're trying to edit hematopoietic stem cells, you can take them out of the body. Once they're out of the body, you have many more methods you can use to deliver efficiently into them. You can electroporated messenger, RNA or even ribonuclear proteins. You can treat with lipids or viruses, you can edit and then put them back into the body. But as you already mentioned, that's sort of a unique feature of blood cells that isn't applicable to the heart or the brain, for example, or the eyes. So then that brings us to viral vectors. There are a variety of clinically validated viral methods for delivery. AAV— adeno associated virus— is probably the most diverse, most relevant, and one of the best tolerated viral delivery methods. The beauty of AAV is that it can deliver to a variety of tissues. AAV can deliver into spinal cord neurons, for example, into retinal cells, into the heart, into the liver, into a few other tissues as well.
(28:48):
And that diversity of being able to choose AAV capsids that are known to get into the types of tissues that you're trying to target is a great strength of that approach. One of the downsides of AAV for gene editing agents is that their delivery tends to be fairly durable. You can engineer AAVs into next generation capsids that sort of get rid of themselves or the gene editing agents get rid of themselves. But classic AAV tends to stay around in patients for a long time, at least months, for example, and possibly years. And we also don't yet have a good way, clinically validated way of re-dosing AAV. And once you administer high doses of AAV in a patient that tends to provoke high-titer, neutralizing antibodies against those AAVs making it difficult to then come back six months or a year later and dose again with an AAV.
(29:57):
So researchers are on the bright side, have become very good at engineering and evolving in the laboratory next generation AAVs that can go to greater diversity issues that can be more potent. Potency is important because if you can back off the dose, maybe you can get around some of these immunogenicity issues. And I think we will see a renaissance with AAV that will further broaden its clinical scope. Even though I appreciate that the decisions by a couple large pharma companies to sort of pull out of using AAV for gene therapy seemed to cause people to, I think prematurely conclude that AAV has fallen out of favor. I think for gene therapy, it's quite different than gene editing. Gene therapy, meaning you are delivering a healthy copy of the gene, and you need to keep that healthy copy of the gene in the patient for the rest of the patient's life.
(30:59):
That's quite different than gene editing where you just need the edit to take place over days to weeks, and then you want the editing agent to actually go away and you never want to come back. I think AAV will used to deliver gene editing agents will avoid some of the clinical challenges like how do we redose? Because you shouldn't need to redose if the gene editing clinical trial proceeds as you hope. And then you mentioned these virus-like particles. So we became interested in virus-like particles as other labs have because they offer some of the best strengths of non-viral and viral approaches like non-viral approaches such as LMPs. They deliver the transient form of a gene editing agent. In fact, they can deliver the fully assembled protein RNA complex of a base editor or a prime editor or a CRISPR nuclease. So in its final form, and that means the exposure of the cell to the editing agent is minimized.
(32:15):
You can treat with these virus-like particles, deliver the protein form of these gene editing agents, allow the on-target site to get edited. And then since the half-life of these proteins tends to be very small, roughly 24 hours for example, by a week later, there should be very little of the material left in the animal or prospectively in the patient virus-like particles, as you call them, neutered viruses, they lack viral DNA or RNA. They don't have the ability to integrate a virus's genome into the human genome, which can cause some undesired consequences. They don't randomly introduce DNA into our genomes, therefore, and they disappear more transiently than viruses like AAV or adenoviruses or other kinds of lentiviruses that have been used in the clinic. So these virus-like particles or VLP offer really some of the best strengths on paper at least of both viral and non-viral delivery.
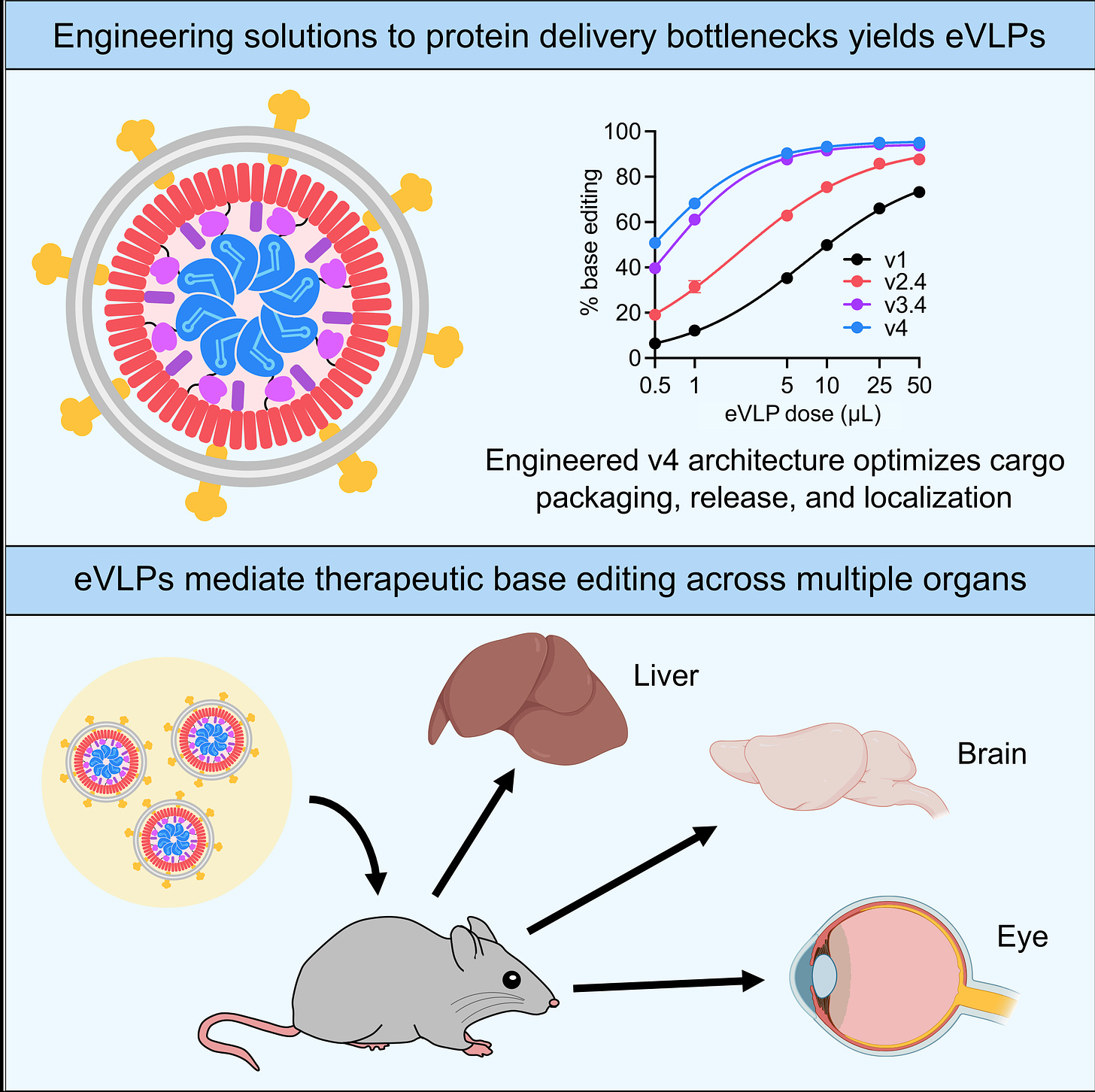
(33:30):
Their limitation thus far has been that there really haven't been examples of potent in vivo delivery of cargoes like gene editing agents using virus-like particles. And so we recently set out to figure out why, and we identified several bottlenecks, molecular bottlenecks that seemed to be standing in the way of virus-like particles, doing a much more efficient job at delivering inside of an animal. (Figure from that paper below.) And we engineered solutions to each of these first three molecular bottlenecks, and we've identified a couple more since. And that resulted in what we call VLPs engineered virus-like particles. And as you pointed out, Keith Joung and myself, co-founded a company called Nvelop to try to bring these technologies and other kinds of molecular delivery technologies, next generation delivery technologies to patients.
Eric Topol (34:28):
Well, that gets me to the near wrapping up, and that is the almost imagination you could use about where all this can go in the future. Recently, I spoke to a mutual friend Fyodor Urnov, who talked about wouldn't it be amazing if for people with chronic pain you could just genome edit neurons their spinal cord? As you already touched on recently, Jennifer Doudna, who we both know talked about editing to prevent Alzheimer's disease. Well, that may be a little far off in time, but at least people are talking about these things that is not, we're not talking about germline editing, we're just talking about somatic cell and being able to approach conditions that have previously been either unapproachable or of limited success and potential of curing. So this field continues to evolve and you and all your colleagues are a big part of how this has evolved as quickly as it has. What are your thoughts about, are there any bounds to the potential in the longer term for genome editing? Right.
David Liu (35:42):
It's a great question because all of the early uses of gene editing in people are appropriately focused on people who are at dire risk of having shorter lives or very poor quality of life as it should be for a new kind of therapeutic because the risks are high until we continue to validate the clinical benefit of these gene editing treatments. And therefore we want to choose patients the highest that face the poorest prognosis where the risk benefit ratio favors treatment as strongly as possible. But your question, I think very accurately highlights that our genome and changes to it determine far more than whether you have a serious genetic disorder like Sickle Cell Disease or Progeria or Cystic Fibrosis or Familial Hypercholesterolemia or Tay-Sachs disease. And being able to not just correct mutations that are associated with devastating genetic disorders, but perhaps take control of our genomes in more sophisticated way that you pointed out two examples that I think are very thought provoking to treat chronic pain permanently to lower the risk of horrible diseases that affect so many families devastating to economies worldwide as well, like Alzheimer's disease, Parkinson's disease, the genetic risk factors that are the strongest genetic determinants of diseases like Alzheimer's disease are actually, there are several that are known already.
(37:36):
And an interesting possibility for the future, it isn't going to happen in the next few years, but it might happen within the next 10 or 20 years, might be to use gene editing to precisely change some of those most grievous alleles that are risk factors for Alzheimer's disease like a apoE4, to change them to the genetic forms that have normal or even reduced risk for Alzheimer's disease. That's a very tough clinical trial to run, but I'd say not any tougher than the dozens of most predominantly failed Alzheimer's clinical trials that have probably collectively accounted for hundreds of billions of dollars of investment
Eric Topol (38:28):
Easily.
David Liu (38:31):
And all of that speaks to the fact that Alzheimer's disease, for example, is enormous burden on society by every measure. So it's worth investing and major resources and taking major risks to try to create perhaps preventative treatments that just lower our risk globally. Getting there will require that these pioneering early clinical trials for gene editing are smashing successes. I'm optimistic that they will be, there will be bumps in the road because there always are bumps in the road. There will be patients who have downturns in their health and everyone will wonder whether those patients had a downturn because of a gene editing treatment they received. And ascertaining whether that's the case will be very important. But as these trials continue to progress, and as they continue hopefully on this quite positive trajectory to date, it's tempting to imagine a future where we can use precise gene editing methods. For example, you can install a variety using prime editing, a variety of alleles that naturally occur in people that reduce the risk of Alzheimer's disease or Parkinson's disease like the mutation that 0.1% of Icelandic people and almost nobody else has in amyloid precursor protein changing alanine 673 to threonine (A673T).
(40:09):
It is very thought provoking, and I don't think society is ready now to take that step, but I think if things continue to proceed on this promising trajectory, it's inevitable because arguably, the defining trait of our species is that we use every ounce of our talents and our gifts and our resources and our creativity to try to improve our lives and those of our children. And I don't think if we have ways of treating genetic diseases or even of reducing grievous genetic disease risk, that we will be able to sit on our hands and not take steps towards that kind of future solon as those technologies continue to be validated in the clinic as being safe and efficacious. It's, I teach a gene editing class and I walk them through a slippery slope at the end of five ethics cases, starting with progeria, where most people would say having a single C of T mutation in one gene that you, by definition didn't inherit from mom or dad.
(41:17):
It just happened spontaneously. That gives you an average lifespan of 14 and a half years and strongly affects other aspects of the quality of your life and your family's life that if you can change as we did in animals that T back into a C and correct the disease and rescue many of the phenotypes and extend lifespan, that that's an ethical use of gene editing. Treating genetic deafness is the second case. It's a little bit more complicated because many people in the deaf community don't view deafness as a disability. It's at least a more subjective situation than progeria. But then there are other cases like changing apoE4 to apoE3 or even apoE2 with the lower than normal risk of Alzheimer's disease, or installing that Icelandic mutation and amyloid precursor protein that substantially lowers risk of Alzheimer's disease. And then finally, you can, I always provoke a healthy debate in the class at the end by pointing out that in the 1960s, one of the long distance cross country alpine skiing records was set by a man who had a naturally occurring mutation in his EPO receptor, his erythropoietin receptor, so that his body always thought he was on EPO as if he were dosing on EPO, although that was of course before the era of EPO dosing was really possible, but it was just a naturally occurring mutation in this case, in his family.
(42:48):
And when I first started teaching this class, most students could accept using gene editing to treat progeria, but very few were willing to go even past that, even to genetic deafness, certainly not to changing a ApoE risk factors for Alzheimer's. Nowadays, I'd say the 50% vote point is somewhere between case three and case four, most people are actually say, yeah, especially since they have family members who've been through Alzheimer's disease. If they are a apoE4, some of them are a apoE4/apoE4 [homozygotes], why not change that to a apoE3 or even an ApoE2 or as one student challenged the class this year, if you were born with a apoE2, would you want to change it to a ApoE3 so you could be more normal? Most people would say, no, there's no way I would do that.
(43:49):
And for the first time this year, there were one or two students who actually even defended the idea of putting in a mutation in erythropoietin receptor to increased increase their endurance under low oxygen conditions. Of course, it's also presumably useful if you ever, God forbid, are treated with a cancer chemotherapeutic. Normally you get erythropoietin to try to restore some, treat some of the anemia that can result, and this student was making a case, well, why wouldn't we? If this is a naturally occurring mutation that's been shown to benefit certain people doing certain things. I don't think that's a general societal view. And I am a little bit skeptical we'll ever get widespread acceptance of case number five. But I think all of it is healthy stimulates a healthy discussion around the surprisingly gentle continuum between disease treatment, disease prevention, and what some would call human improvement.And it used to be that even the word human improvement was sort of an anathema. I think now at least the students in my class are starting to rethink what does that really mean? We improving ourselves a number of ways genetically and otherwise by virtue of our lifestyles, by virtue of who we choose to procreate with. So it's a really interesting debate, and I think the rapid development and now clinical progression and now approval, regulatory approval of gene editing drugs will play a central role in this discussion.
Eric Topol (45:38):
No question. I mean, also just to touch on the switch from a apoE4 to apoE2, you would get a potential 2-fer of lesser risk for Alzheimer's and a longer lifespan. So I mean, there's a lot of things here. The thing that got me years ago, I mean, this is many years ago at a meeting with George Church and he says, we're going to just edit 60 genes and then we can do all sorts of xeno-pig transplants and forget the problem of donors. And it's happening now.
David Liu (46:11):
Yeah, I mean, he used a base editor to edit hundreds of genes at once, if not thousands of
Eric Topol (46:16):
That's why it's just, yeah, no, it's just extraordinary. And I think people need to be aware that opportunities here, as you say, with potential bumps along the way, unquestionably, is almost limitless. So this has been a masterclass thanks to you, David, in where we are, where we're headed in genome editing at a very extraordinary time where we've really seeing things click. And I just want to also add that you're going to be here with a conference in La Jolla in January, I think, on base and prime editing. Is that right? So for those who are listeners who are into this topic, maybe they can also hear the latest, I'm sure there'll be more between now and next. Well, several weeks from now at your, it's a
David Liu (47:12):
Conference on, it's the fifth international conference on base and prime editing and associated enzymes, the somewhat baroque name. And I will at least be giving a virtual talk there. It actually overlaps with the talk I'm giving at Rockefeller that time. Ah, okay, cool. But I'm speaking at the conference either in person or virtually.
Eric Topol (47:34):
Yeah. Well, anytime we get to hear from you and the field, of course it's enlightening. So thanks so much for joining. Thank you
David Liu (47:42):
For having me. And thank you also for all of your, I think, really important public service in connecting appropriately the ground truths about science and vaccines and other things to people. I think that's very much appreciated by scientists like myself.
Eric Topol (48:00):
Oh, thanks David.
Thanks for listening, reading, and subscribing to Ground Truths.
To be clear, this is a hybrid format, roughly alternating between analytical newsletters/essays and podcasts with exceptional people, attempting to achieve about 2 posts per week. It’s all related to cutting-edge advances in life science, medicine, and information tech (A.I.)
All content is free. If you wish to become a paid subscriber know that all proceeds go to Scripps Research.














Share this post